6WLC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-19 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
8EBG
 
 | | Crystal structure of the probable FhuD FeIII-dicitrate-binding domain protein FecB from Mycobacterium tuberculosis | | 分子名称: | ACETIC ACID, FEIII-dicitrate-binding periplasmic lipoprotein FecB, FORMIC ACID, ... | | 著者 | Cuff, M, Kim, Y, Endres, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2022-08-31 | | 公開日 | 2022-09-14 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | Crystal structure of the probable FhuD FeIII-dicitrate-binding domain protein FecB from Mycobacterium tuberculosis
To Be Published
|
|
5BMZ
 
 | | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA. | | 分子名称: | DNA (5'-D(P*GP*AP*AP*TP*AP*TP*CP*AP*GP*TP*TP*AP*AP*AP*CP*TP*GP*AP*TP*AP*TP*TP*C)-3'), HcaR protein | | 著者 | Kim, Y, Joachimiak, G, Biglow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-05-25 | | 公開日 | 2015-10-14 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (3.001 Å) | | 主引用文献 | Crystal Structure of Putative MarR Family Transcriptional Regulator HcaR from Acinetobacter sp. ADP complexed with 24mer DNA.
To Be Published
|
|
8DIL
 
 | | Crystal structure of putative nitroreductase from Salmonella enterica | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Chang, C, Skarina, T, Mesa, N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-06-29 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of putative nitroreductase from Salmonella enterica
to be published
|
|
8EP6
 
 | | Crystal Structure of the Beta-lactamase Class D from Chitinophaga pinensis in complex with Avibactam | | 分子名称: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, ACETIC ACID, Beta-lactamase Class D Cpin_0907 | | 著者 | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-10-05 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-02-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of the Beta-lactamase Class D from Chitinophaga pinensis in the complex with Avibactam.
To Be Published
|
|
8EP7
 
 | | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in complex with NADP | | 分子名称: | ACETIC ACID, Ketol-acid reductoisomerase (NADP(+)) 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Kim, Y, Maltseva, N, Osipiuk, J, Gu, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-10-05 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of the Ketol-acid Reductoisomerase from Bacillus anthracis in the complex with NADP.
To Be Published
|
|
8EBC
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP | | 分子名称: | FORMIC ACID, GLYCEROL, INOSINIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Osipiuk, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2022-08-31 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP
To Be Published
|
|
6MHM
 
 | | Crystal structure of human acid ceramidase in covalent complex with carmofur | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Dementiev, A, Joachimiak, A, Doan, N. | | 登録日 | 2018-09-18 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.743 Å) | | 主引用文献 | Molecular Mechanism of Inhibition of Acid Ceramidase by Carmofur.
J. Med. Chem., 62, 2019
|
|
8EY4
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37 | | 分子名称: | Cys_rich_CPCC domain-containing protein, FE (III) ION, PT-VENN domain-containing protein | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-10-26 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37
To Be Published
|
|
8EY3
 
 | | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37 | | 分子名称: | Cys_rich_CPCC domain-containing protein, FE (III) ION, SODIUM ION | | 著者 | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-10-26 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37
To Be Published
|
|
5C0P
 
 | | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Endo-arabinase, ... | | 著者 | Tan, K, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-06-12 | | 公開日 | 2015-07-01 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.532 Å) | | 主引用文献 | The crystal structure of endo-arabinase from Bacteroides thetaiotaomicron VPI-5482
To Be Published
|
|
6N7F
 
 | | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD. | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BICARBONATE ION, ... | | 著者 | Minasov, G, Shuvalova, L, Shabalin, I.G, Grabowski, M, Olphie, A, Cardona-Correa, A, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-11-27 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 1.90 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus pyogenes in Complex with FAD.
To Be Published
|
|
5E2F
 
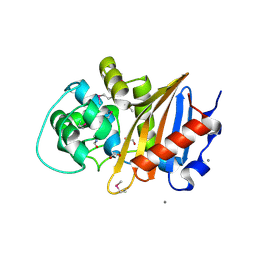 | | Crystal Structure of Beta-lactamase class D from Bacillus subtilis | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase YbxI, CALCIUM ION | | 著者 | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-01 | | 公開日 | 2015-10-14 | | 最終更新日 | 2022-03-30 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystal Structure of Beta-lactamase class D from Bacillus subtilis
To Be Published
|
|
1I36
 
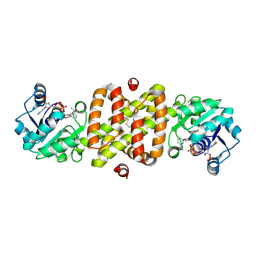 | | Structure of Conserved Protein MTH1747 of Unknown Function Reveals Structural Similarity with 3-Hydroxyacid Dehydrogenases | | 分子名称: | CONSERVED HYPOTHETICAL PROTEIN MTH1747, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Korolev, S.V, Dementieva, I.S, Christendat, D, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2001-02-13 | | 公開日 | 2002-05-15 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | STRUCTURAL SIMILARITIES OF MTH1747 HYPOTHETICAL PROTEIN FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH 3-HYDROXYACID DEHYDROGENASES
to be published
|
|
5E2G
 
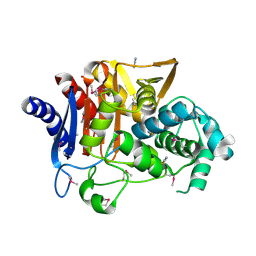 | | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia | | 分子名称: | ACETIC ACID, Beta-lactamase, THIOCYANATE ION | | 著者 | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-01 | | 公開日 | 2015-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.651 Å) | | 主引用文献 | Crystal Structure of D-alanine Carboxypeptidase AmpC from Burkholderia cenocepacia
To Be Published
|
|
5E2E
 
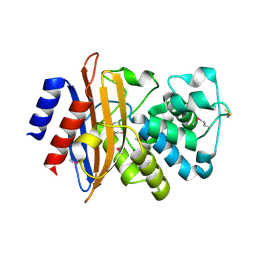 | | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica | | 分子名称: | Beta-lactamase | | 著者 | Kim, Y, Joachimiak, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-01 | | 公開日 | 2015-10-28 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Beta-lactamase Precursor BlaA from Yersinia enterocolitica
To Be Published
|
|
5E3E
 
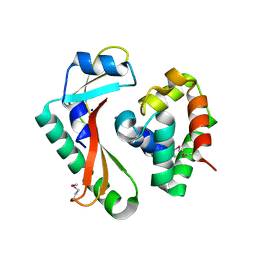 | | Crystal structure of CdiA-CT/CdiI complex from Y. kristensenii 33638 | | 分子名称: | CdiI immunity protein, Large exoprotein involved in heme utilization or adhesion, SODIUM ION | | 著者 | Michalska, K, Joachimiak, G, Jedrzejczak, R, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-02 | | 公開日 | 2015-11-25 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily.
Nucleic Acids Res., 45, 2017
|
|
1GRL
 
 | | THE CRYSTAL STRUCTURE OF THE BACTERIAL CHAPERONIN GROEL AT 2.8 ANGSTROMS | | 分子名称: | GROEL (HSP60 CLASS) | | 著者 | Braig, K, Otwinowski, Z, Hegde, R, Boisvert, D.C, Joachimiak, A, Horwich, A.L, Sigler, P.B. | | 登録日 | 1995-03-07 | | 公開日 | 1995-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of the bacterial chaperonin GroEL at 2.8 A.
Nature, 371, 1994
|
|
7L91
 
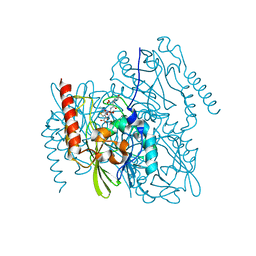 | | Structure of Metallo Beta-Lactamase L1 in a Complex with Hydrolyzed Moxalactam Determined by Pink-Beam Serial Crystallography | | 分子名称: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | 著者 | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Vukica, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-01-01 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
4WRP
 
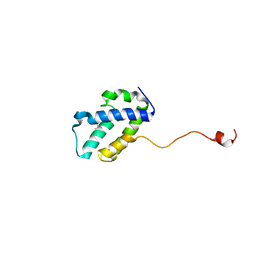 | | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | 分子名称: | Uncharacterized protein | | 著者 | Cuff, M.E, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-10-24 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The C-terminal domain of gene product lpg0944 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1.
To Be Published
|
|
1P99
 
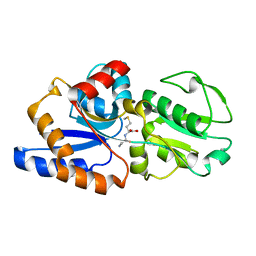 | | 1.7A crystal structure of protein PG110 from Staphylococcus aureus | | 分子名称: | GLYCINE, Hypothetical protein PG110, METHIONINE | | 著者 | Zhang, R, Zhou, M, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2003-05-09 | | 公開日 | 2004-01-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions.
Biochemistry, 43, 2004
|
|
7UHM
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (150 ms Snapshot) | | 分子名称: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | 著者 | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-03-27 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
7UHO
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (500 ms Snapshot) | | 分子名称: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | 著者 | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-03-27 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
1D9K
 
 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | 分子名称: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | 著者 | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | 登録日 | 1999-10-28 | | 公開日 | 1999-12-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
7UHJ
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Non-Hydrolyzed Moxalactam (60 ms Snapshot) | | 分子名称: | (1R,6R,7R)-7-[(2R)-2-carboxypropanamido]-7-methoxy-3-methyl-8-oxo-5-oxa-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | 著者 | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-03-27 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
