7Z1B
 
 | | Nanobody H11-A10 and F2 bound to RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A10, Nanobody F2, ... | | 著者 | Mikolajek, H, Naismith, J.H. | | 登録日 | 2022-02-24 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2W82
 
 | | The structure of ArdA | | 分子名称: | ORF18 | | 著者 | McMahon, S.A, Roberts, G.A, Carter, L.G, Cooper, L.P, Liu, H, White, J.H, Johnson, K.A, Sanghvi, B, Oke, M, Walkinshaw, M.D, Blakely, G, Naismith, J.H, Dryden, D.T.F. | | 登録日 | 2009-01-08 | | 公開日 | 2009-01-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Extensive DNA Mimicry by the Arda Anti-Restriction Protein and its Role in the Spread of Antibiotic Resistance.
Nucleic Acids Res., 37, 2009
|
|
7Z7X
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-H6 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H6, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9R
 
 | |
7Z86
 
 | |
7Z6V
 
 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11, ... | | 著者 | Weckener, M, Naismith, J.H, Vogirala, V.K. | | 登録日 | 2022-03-14 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZB2
 
 | | apo macrocyclase OphP | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZAZ
 
 | | macrocyclase OphP with ZPP | | 分子名称: | 1,2-ETHANEDIOL, BICARBONATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZB1
 
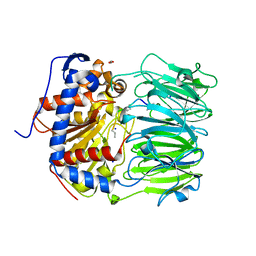 | | S580A with 18mer | | 分子名称: | 1,2-ETHANEDIOL, 18mer, BICARBONATE ION, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7ZB0
 
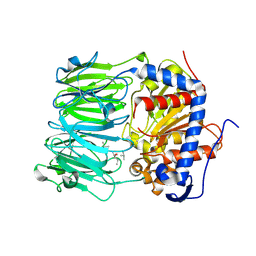 | | macrocyclase OphP with 15mer | | 分子名称: | 1,2-ETHANEDIOL, 15mer, BICARBONATE ION, ... | | 著者 | Song, H, Naismith, J.H. | | 登録日 | 2022-03-23 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Biorxiv, 2022
|
|
7Z85
 
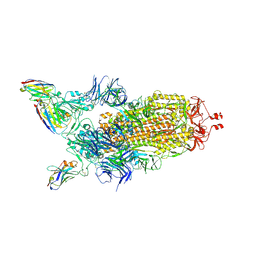 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-B5 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-B5, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-16 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z9Q
 
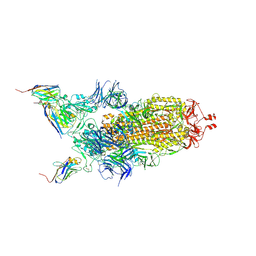 | | CRYO-EM STRUCTURE OF SARS-COV-2 SPIKE : H11-A10 nanobody complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-A10, ... | | 著者 | Weckener, M, Naismith, J.H. | | 登録日 | 2022-03-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8AP0
 
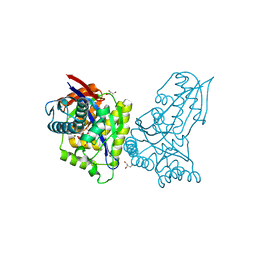 | | ForT Mutant T138V | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 4-azanyl-1~{H}-pyrazole-3,5-dicarboxylic acid, Beta-ribofuranosylaminobenzene 5'-phosphate synthase, ... | | 著者 | Li, W, Naismith, J.H. | | 登録日 | 2022-08-09 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase.
Open Biology, 13, 2023
|
|
8AOZ
 
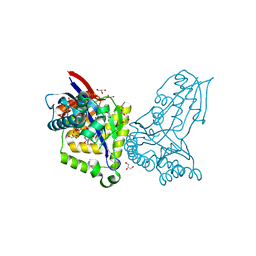 | | ForT Mutant L24A | | 分子名称: | 1,2-ETHANEDIOL, 4-azanyl-3-[(2~{S},3~{R},4~{S},5~{R})-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]-1~{H}-pyrazole-5-carboxylic acid, Beta-ribofuranosylaminobenzene 5'-phosphate synthase, ... | | 著者 | Li, W, Naismith, J.H. | | 登録日 | 2022-08-09 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Experimental and computational snapshots of C-C bond formation in a C-nucleoside synthase.
Open Biology, 13, 2023
|
|
8ANX
 
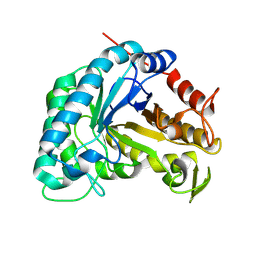 | |
8AG9
 
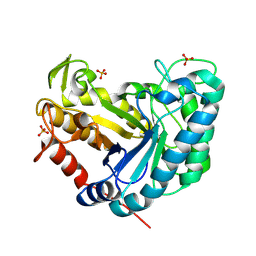 | |
1PM7
 
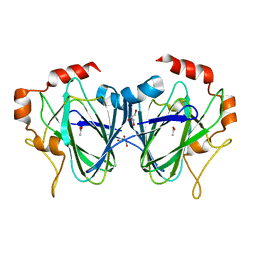 | |
2X5F
 
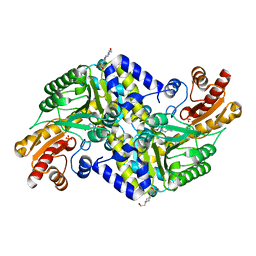 | | Crystal structure of the methicillin-resistant Staphylococcus aureus Sar2028, an aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate dependent aminotransferase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASPARTATE_TYROSINE_PHENYLALANINE PYRIDOXAL-5' PHOSPHATE-DEPENDENT AMINOTRANSFERASE, MAGNESIUM ION, ... | | 著者 | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | 登録日 | 2010-02-08 | | 公開日 | 2010-07-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
2X3F
 
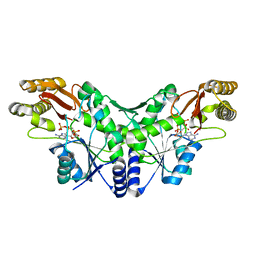 | | Crystal Structure of the Methicillin-Resistant Staphylococcus aureus Sar2676, a Pantothenate Synthetase. | | 分子名称: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PANTHOTHENATE SYNTHETASE, SULFATE ION | | 著者 | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | 登録日 | 2010-01-24 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
1NYW
 
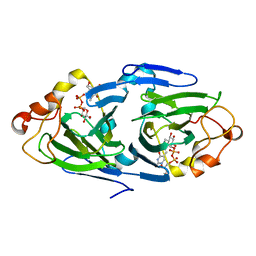 | | The high resolution structures of RmlC from Streptoccus suis in complex with dTDP-D-glucose | | 分子名称: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | 著者 | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | 登録日 | 2003-02-14 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NZC
 
 | | The high resolution structures of RmlC from Streptococcus suis in complex with dTDP-D-xylose | | 分子名称: | NICKEL (II) ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | 著者 | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | 登録日 | 2003-02-17 | | 公開日 | 2003-06-24 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1NXM
 
 | | The high resolution structures of RmlC from Streptococcus suis | | 分子名称: | dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | 著者 | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | 登録日 | 2003-02-11 | | 公開日 | 2003-06-24 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
1OC2
 
 | |
1OFN
 
 | | Purification, crystallisation and preliminary structural studies of dTDP-4-keto-6-deoxy-glucose-5-epimerase (EvaD) from Amycolatopsis orientalis; the fourth enzyme in the dTDP-L-epivancosamine biosynthetic pathway. | | 分子名称: | GLYCEROL, PCZA361.16 | | 著者 | Merkel, A.B, Naismith, J.H. | | 登録日 | 2003-04-17 | | 公開日 | 2004-04-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Purification, Crystallization and Preliminary Structural Studies of Dtdp-4-Keto-6-Deoxy-Glucose-5-Epimerase (Evad) from Amycolatopsis Orientalis, the Fourth Enzyme in the Dtdp-L-Epivancosamine Biosynthetic Pathway.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7BHJ
 
 | |
