5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | 分子名称: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.502 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSJ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C delta-HEAD2 COMPLEX | | 分子名称: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | 分子名称: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
6XRT
 
 | |
4Y94
 
 | |
4Y93
 
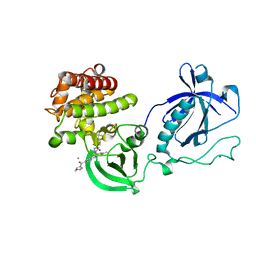 | | Crystal structure of the PH-TH-kinase construct of Bruton's tyrosine kinase (Btk) | | 分子名称: | 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, CALCIUM ION, Non-specific protein-tyrosine kinase,Non-specific protein-tyrosine kinase, ... | | 著者 | Wang, Q, Kuriyan, J. | | 登録日 | 2015-02-16 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.695 Å) | | 主引用文献 | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
4Y95
 
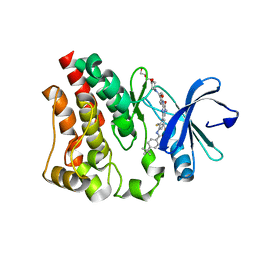 | | Crystal structure of the kinase domain of Bruton's tyrosine kinase with mutations in the activation loop | | 分子名称: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-tert-butyl-N-[2-methyl-3-(4-methyl-6-{[4-(morpholin-4-ylcarbonyl)phenyl]amino}-5-oxo-4,5-dihydropyrazin-2-yl)phenyl]benzamide, BETA-MERCAPTOETHANOL, ... | | 著者 | Wang, Q, Rosen, C.E, Kuriyan, J. | | 登録日 | 2015-02-16 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate.
Elife, 4, 2015
|
|
2R63
 
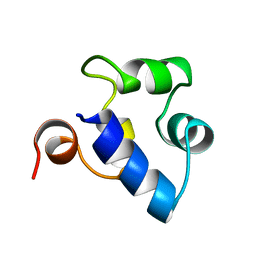 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-13 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
5MU3
 
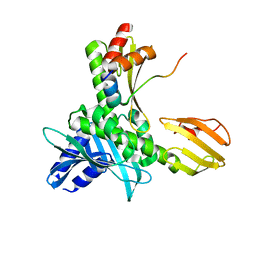 | |
1LCK
 
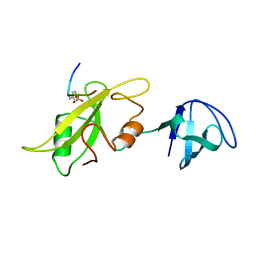 | |
1LCJ
 
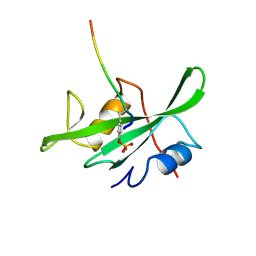 | |
2GUM
 
 | |
2LCK
 
 | |
2O6G
 
 | |
7QRE
 
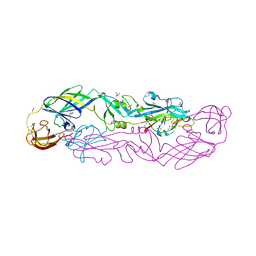 | | Structure of the hetero-tetramer complex between precursor membrane protein fragment (pr) and envelope protein (E) from tick-borne encephalitis virus | | 分子名称: | ACETATE ION, Envelope protein E, Genome polyprotein, ... | | 著者 | Vaney, M.C, Dellarole, M, Rey, F.A. | | 登録日 | 2022-01-11 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
7QRF
 
 | | Structure of the dimeric complex between precursor membrane ectodomain (prM) and envelope protein ectodomain (E) from tick-borne encephalitis virus | | 分子名称: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Envelope protein E, ... | | 著者 | Vaney, M.C, Rouvinski, A, Rey, F.A. | | 登録日 | 2022-01-11 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Evolution and activation mechanism of the flavivirus class II membrane-fusion machinery.
Nat Commun, 13, 2022
|
|
2O61
 
 | |
7MGS
 
 | |
7MGR
 
 | |
1PRA
 
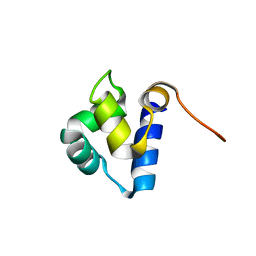 | |
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-08 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1TZN
 
 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore bound to the VWA domain of CMG2, an anthrax toxin receptor | | 分子名称: | Anthrax toxin receptor 2, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | 登録日 | 2004-07-10 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TZO
 
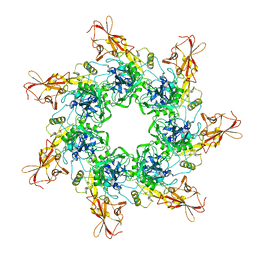 | | Crystal Structure of the Anthrax Toxin Protective Antigen Heptameric Prepore | | 分子名称: | CALCIUM ION, Protective antigen | | 著者 | Lacy, D.B, Wigelsworth, D.J, Melnyk, R.A, Collier, R.J. | | 登録日 | 2004-07-10 | | 公開日 | 2004-08-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structure of heptameric protective antigen bound to an anthrax toxin receptor: A role for receptor in pH-dependent pore formation
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
5UKN
 
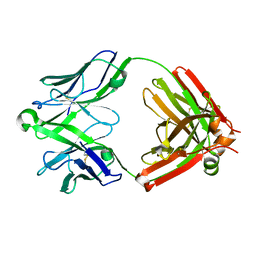 | |
5UKP
 
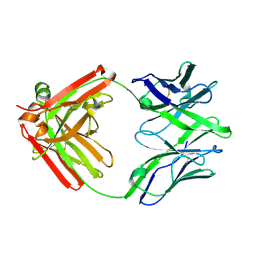 | |
