5KNZ
 
 | | Human Islet Amyloid Polypeptide Segment 19-SGNNFGAILSS-29 with Early Onset S20G Mutation Determined by MicroED | | 分子名称: | hIAPP(residues 19-29)S20G | | 著者 | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | 登録日 | 2016-06-28 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | 主引用文献 | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
5HQX
 
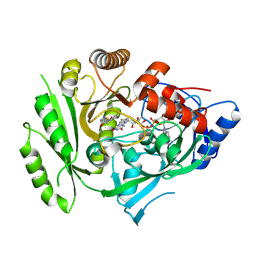 | | Crystal structure of maize cytokinin oxidase/dehydrogenase 4 (ZmCKO4) in complex with phenylurea inhibitor HETDZ | | 分子名称: | 1-[2-(2-hydroxyethyl)phenyl]-3-(1,2,3-thiadiazol-5-yl)urea, Cytokinin dehydrogenase 4, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Kopecny, D, Koncitikova, R, Briozzo, P. | | 登録日 | 2016-01-22 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Novel thidiazuron-derived inhibitors of cytokinin oxidase/dehydrogenase.
Plant Mol.Biol., 92, 2016
|
|
1CP7
 
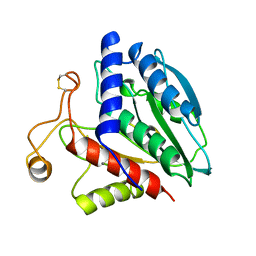 | | AMINOPEPTIDASE FROM STREPTOMYCES GRISEUS | | 分子名称: | AMINOPEPTIDASE, CALCIUM ION, ZINC ION | | 著者 | Gilboa, R, Greenblatt, H.M, Perach, M, Spungin-Bialik, A, Lessel, U, Schomburg, D, Blumberg, S, Shoham, G. | | 登録日 | 1999-06-10 | | 公開日 | 2000-05-03 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Interactions of Streptomyces griseus aminopeptidase with a methionine product analogue: a structural study at 1.53 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
9CO2
 
 | |
5HNR
 
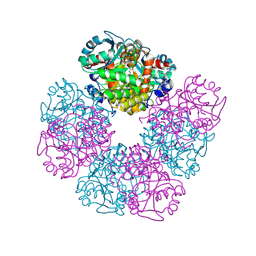 | | The X-ray structure of octameric human native 5-aminolaevulinic acid dehydratase. | | 分子名称: | DELTA-AMINO VALERIC ACID, Delta-aminolevulinic acid dehydratase, SULFATE ION, ... | | 著者 | Mills-Davies, N.L, Thompson, D, Shoolingin-Jordan, P.M, Erskine, P.T, Cooper, J.B. | | 登録日 | 2016-01-18 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structural studies of substrate and product complexes of 5-aminolaevulinic acid dehydratase from humans, Escherichia coli and the hyperthermophile Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1CSB
 
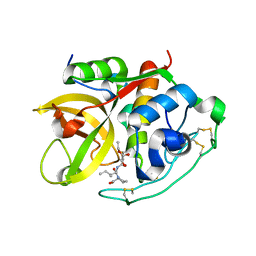 | |
9GCF
 
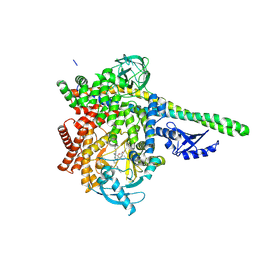 | | HUMAN PI3KDELTA IN COMPLEX WITH ISOCUMARIN INHIBITOR CHF-6523 | | 分子名称: | 3-[(1S)-1-[4-azanyl-3-(5-oxidanylpyridin-3-yl)pyrazolo[3,4-d]pyrimidin-1-yl]ethyl]-4-[3-[(4-methylpiperazin-1-yl)methyl]phenyl]isochromen-1-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | 著者 | Pala, D, Bruno, P, Capelli, A.M, Biagetti, M. | | 登録日 | 2024-08-01 | | 公開日 | 2024-12-18 | | 最終更新日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Discovery of CHF-6523, an Inhaled Selective PI3K delta Inhibitor for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 68, 2025
|
|
8RNZ
 
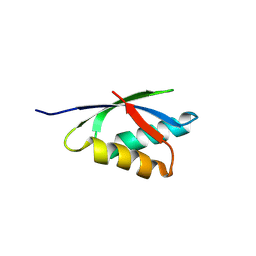 | |
9GDI
 
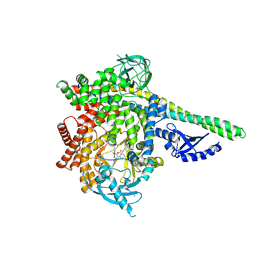 | | HUMAN PI3KDELTA IN COMPLEX WITH ISOCUMARIN INHIBITOR 10 | | 分子名称: | 3-[(1S)-1-[4-azanyl-3-(3-fluoranyl-5-oxidanyl-phenyl)pyrazolo[3,4-d]pyrimidin-1-yl]ethyl]-4-(1-methyl-3,6-dihydro-2H-pyridin-4-yl)isochromen-1-one, CHLORIDE ION, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | 著者 | Pala, D, Bruno, P, Capelli, A.M, Biagetti, M. | | 登録日 | 2024-08-05 | | 公開日 | 2024-12-18 | | 最終更新日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Discovery of CHF-6523, an Inhaled Selective PI3K delta Inhibitor for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 68, 2025
|
|
5AOU
 
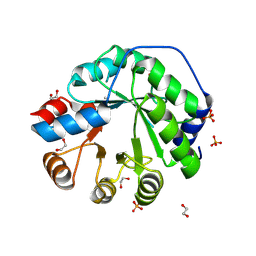 | | Structure of the engineered retro-aldolase RA95.5-8F apo | | 分子名称: | 1,2-ETHANEDIOL, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE, PHOSPHATE ION | | 著者 | Obexer, R, Mittl, P, Hilvert, D. | | 登録日 | 2015-09-11 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase.
Nat Chem, 9, 2017
|
|
9GG9
 
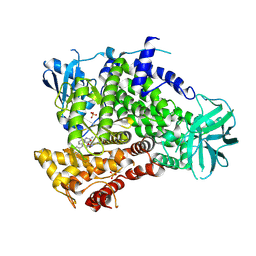 | | HUMAN PI3KGAMMA IN COMPLEX WITH ISOCUMARIN INHIBITOR 11 | | 分子名称: | 3-[(1~{S})-1-[4-azanyl-3-(3-fluoranyl-5-oxidanyl-phenyl)pyrazolo[3,4-d]pyrimidin-1-yl]ethyl]-4-phenyl-isochromen-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Pala, D, Bruno, P, Capelli, A.M, Biagetti, M. | | 登録日 | 2024-08-13 | | 公開日 | 2024-12-18 | | 最終更新日 | 2025-02-26 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Discovery of CHF-6523, an Inhaled Selective PI3K delta Inhibitor for the Treatment of Chronic Obstructive Pulmonary Disease.
J.Med.Chem., 68, 2025
|
|
1D4K
 
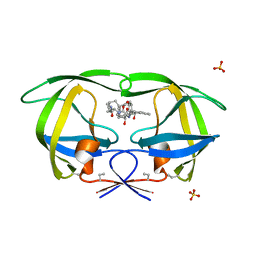 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | 分子名称: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | 著者 | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | 登録日 | 1999-10-04 | | 公開日 | 2000-10-11 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
7O4W
 
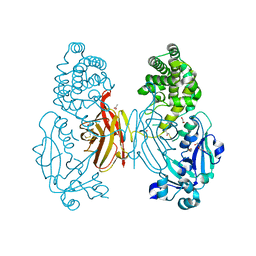 | |
6SHN
 
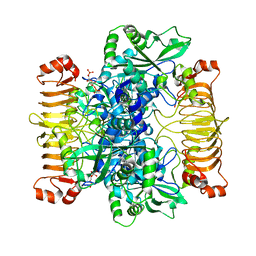 | | Escherichia coli AGPase in complex with FBP. Symmetry C1 | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | 著者 | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | 登録日 | 2019-08-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
9C6O
 
 | |
1P82
 
 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | 分子名称: | 10 kDa chaperonin | | 著者 | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | 登録日 | 2003-05-06 | | 公開日 | 2003-05-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
6U94
 
 | | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344 | | 分子名称: | GLYCEROL, RND efflux system, outer membrane lipoprotein, ... | | 著者 | Horanyi, P.S, Fox III, D, Abendroth, J, Lorimer, D, Edwards, T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2019-09-06 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of RND efflux system, outer membrane lipoprotein, NodT family from Burkholderia mallei ATCC 23344
To Be Published
|
|
9C7Z
 
 | | Hallucinated C3 protein assembly HALC3_919 | | 分子名称: | HALC3_919, TETRAETHYLENE GLYCOL | | 著者 | Ragotte, R, Bera, A, Milles, L.F, Wicky, B.I.M, Baker, D. | | 登録日 | 2024-06-11 | | 公開日 | 2025-05-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Designed miniprotein inhibitors potently inhibit and protect against MERS-CoV
To Be Published
|
|
5IA1
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with MLN8054 | | 分子名称: | 1,2-ETHANEDIOL, 4-{[9-CHLORO-7-(2,6-DIFLUOROPHENYL)-5H-PYRIMIDO[5,4-D][2]BENZAZEPIN-2-YL]AMINO}BENZOIC ACID, Ephrin type-A receptor 2 | | 著者 | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | 登録日 | 2016-02-21 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.036 Å) | | 主引用文献 | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
6GQJ
 
 | | Crystal structure of human c-KIT kinase domain in complex with AZD3229-analogue (compound 18) | | 分子名称: | 2-[4-(6,7-dimethoxyquinazolin-4-yl)oxy-2-methoxy-phenyl]-~{N}-(1-propan-2-ylpyrazol-4-yl)ethanamide, Mast/stem cell growth factor receptor Kit | | 著者 | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | 登録日 | 2018-06-07 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
6V05
 
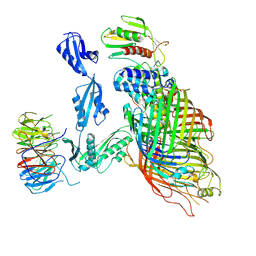 | | Cryo-EM structure of a substrate-engaged Bam complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | 著者 | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | 登録日 | 2019-11-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
5I9V
 
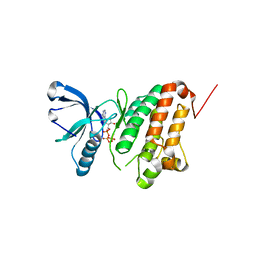 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with AGS | | 分子名称: | Ephrin type-A receptor 2, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | 登録日 | 2016-02-21 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.458 Å) | | 主引用文献 | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5IA3
 
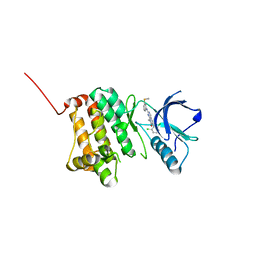 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with PD173955 | | 分子名称: | 6-(2,6-DICHLORO-PHENYL)-8-METHYL-2-(3-METHYLSULFANYL-PHENYLAMINO)-8H-PYRIDO[2,3-D]PYRIMIDIN-7-ONE, Ephrin type-A receptor 2 | | 著者 | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | 登録日 | 2016-02-21 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.788 Å) | | 主引用文献 | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
5ZU1
 
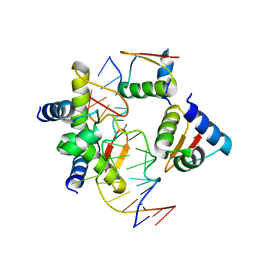 | | Crystal Structure of BZ junction in diverse sequence | | 分子名称: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*AP*GP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*TP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | 著者 | Kim, K.K, Kim, D. | | 登録日 | 2018-05-05 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.009 Å) | | 主引用文献 | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
3EAZ
 
 | |
