3UIU
 
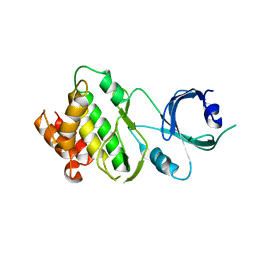 | | Crystal structure of Apo-PKR kinase domain | | 分子名称: | Interferon-induced, double-stranded RNA-activated protein kinase | | 著者 | Li, F, Li, S, Yang, X, Shen, Y, Zhang, T. | | 登録日 | 2011-11-06 | | 公開日 | 2012-11-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.903 Å) | | 主引用文献 | Crystal structure of Apo-PKR kinase domain
TO BE PUBLISHED
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | 分子名称: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | 著者 | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | 登録日 | 2020-07-14 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
3QYC
 
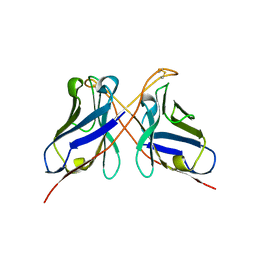 | | Structure of a dimeric anti-HER2 single domain antibody | | 分子名称: | VH domain of IgG molecule | | 著者 | Baral, T.N, Chao, S, Li, S, Tanha, J, Arbabai, M, Wang, S, Zhang, J. | | 登録日 | 2011-03-03 | | 公開日 | 2012-02-08 | | 最終更新日 | 2014-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of a Human Single Domain Antibody Dimer Formed through V(H)-V(H) Non-Covalent Interactions.
Plos One, 7, 2012
|
|
8WO2
 
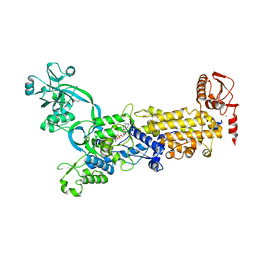 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val-AMP | | 分子名称: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | 著者 | Guo, Y, Li, S, Zhang, T. | | 登録日 | 2023-10-06 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNJ
 
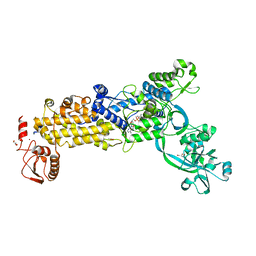 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile-AMP | | 分子名称: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | 著者 | Guo, Y, Li, S, Zhang, T. | | 登録日 | 2023-10-06 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNF
 
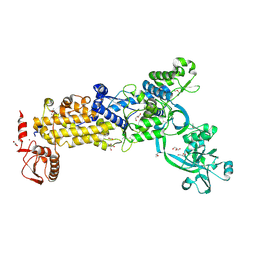 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in apo form | | 分子名称: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | 著者 | Guo, Y, Li, S, Zhang, T. | | 登録日 | 2023-10-05 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WO3
 
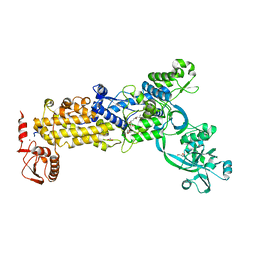 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Mupirocin | | 分子名称: | ACETATE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | 著者 | Guo, Y, Li, S, Zhang, T. | | 登録日 | 2023-10-06 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNI
 
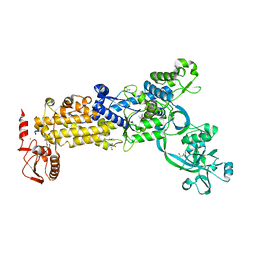 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Val | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | 著者 | Guo, Y, Li, S, Zhang, T. | | 登録日 | 2023-10-06 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8WNG
 
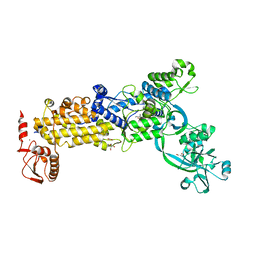 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile | | 分子名称: | ACETATE ION, GLYCEROL, ISOLEUCINE, ... | | 著者 | Guo, Y, Li, S, Zhang, T. | | 登録日 | 2023-10-05 | | 公開日 | 2024-02-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
8GTA
 
 | | Cryo-EM structure of the marine siphophage vB_Dshs-R4C capsid | | 分子名称: | Major capsid protein | | 著者 | Sun, H, Huang, Y, Zheng, Q, Li, S, Zhang, R, Xia, N. | | 登録日 | 2022-09-07 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Structure and proposed DNA delivery mechanism of a marine roseophage.
Nat Commun, 14, 2023
|
|
5DJ5
 
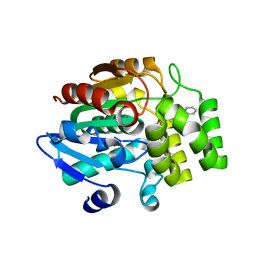 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | 分子名称: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | 著者 | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | 登録日 | 2015-09-01 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | 分子名称: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | 分子名称: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
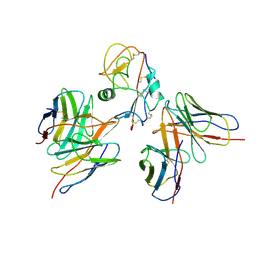 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | 分子名称: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | 著者 | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | 登録日 | 2022-08-02 | | 公開日 | 2024-01-31 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
6AL7
 
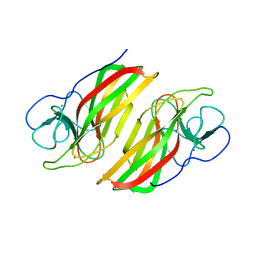 | | Crystal structure HpiC1 F138S | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.687 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL6
 
 | | Crystal structure HpiC1 in P42 space group | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.088 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL8
 
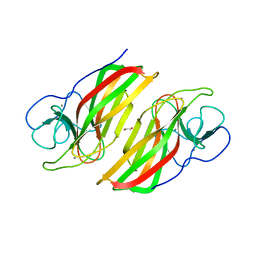 | | Crystal structure HpiC1 Y101F/F138S | | 分子名称: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
4HPH
 
 | | The crystal structure of isomaltulose synthase mutant E295Q from Erwinia rhapontici NX5 in complex with its natural substrate sucrose | | 分子名称: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | 著者 | Xu, Z, Li, S, Xu, H, Zhou, J. | | 登録日 | 2012-10-23 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOW
 
 | | The crystal structure of isomaltulose synthase from Erwinia rhapontici NX5 | | 分子名称: | CALCIUM ION, GLYCEROL, Sucrose isomerase | | 著者 | Xu, Z, Li, S, Xu, H, Zhou, J. | | 登録日 | 2012-10-22 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
4HOZ
 
 | | The crystal structure of isomaltulose synthase mutant D241A from Erwinia rhapontici NX5 in complex with D-glucose | | 分子名称: | CALCIUM ION, GLYCEROL, Sucrose isomerase, ... | | 著者 | Xu, Z, Li, S, Xu, H, Zhou, J. | | 登録日 | 2012-10-23 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Structural Basis of Erwinia rhapontici Isomaltulose Synthase
Plos One, 8, 2013
|
|
1LOH
 
 | | Streptococcus pneumoniae Hyaluronate Lyase in Complex with Hexasaccharide Hyaluronan Substrate | | 分子名称: | Hyaluronate Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Jedrzejas, M.J, Mello, L.V, De Groot, B.L, Li, S. | | 登録日 | 2002-05-06 | | 公開日 | 2002-08-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate.
J.Biol.Chem., 277, 2002
|
|
6UES
 
 | | Apo SAM-IV Riboswitch | | 分子名称: | RNA (119-MER) | | 著者 | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | 登録日 | 2019-09-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6UET
 
 | | SAM-bound SAM-IV riboswitch | | 分子名称: | RNA (119-MER), S-ADENOSYLMETHIONINE | | 著者 | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | 登録日 | 2019-09-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6V5C
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | 分子名称: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | 著者 | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | 登録日 | 2019-12-04 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
1LXK
 
 | | Streptococcus pneumoniae Hyaluronate Lyase in Complex with Tetrasaccharide Hyaluronan Substrate | | 分子名称: | Hyaluronate Lyase, beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Jedrzejas, M.J, Mello, L.V, De Groot, B.L, Li, S. | | 登録日 | 2002-06-05 | | 公開日 | 2002-08-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate.
J.Biol.Chem., 277, 2002
|
|
