6CMX
 
 | | Human Teneurin 2 extra-cellular region | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, alpha-D-mannopyranose, ... | | 著者 | Shalev-Benami, M, Li, J, Sudhof, T, Skiniotis, G, Arac, D. | | 登録日 | 2018-03-06 | | 公開日 | 2018-07-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse.
Cell, 173, 2018
|
|
5HC0
 
 | |
5HC5
 
 | |
8FIA
 
 | | The structure of fly Teneurin self assembly | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Bandekar, S.J, Li, J, Arac, D. | | 登録日 | 2022-12-15 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The structure of fly Teneurin-m reveals an asymmetric self-assembly that allows expansion into zippers.
Embo Rep., 24, 2023
|
|
6LDZ
 
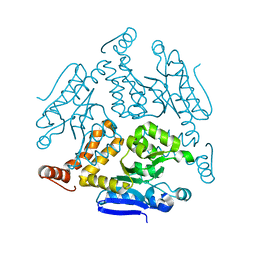 | | Crystal structure of Rv0222 from Mycobacterium tuberculosis | | 分子名称: | Probable enoyl-CoA hydratase EchA1 (Enoyl hydrase) (Unsaturated acyl-CoA hydratase) (Crotonase) | | 著者 | Li, J, Ran, Y.J, Wang, L, Wu, J.H, Ge, B.X, Rao, Z.H. | | 登録日 | 2019-11-23 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Host-mediated ubiquitination of a mycobacterial protein suppresses immunity.
Nature, 577, 2020
|
|
2JOM
 
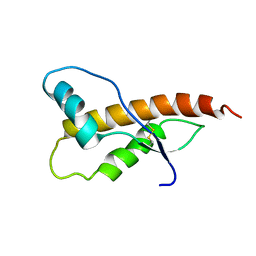 | |
7WNT
 
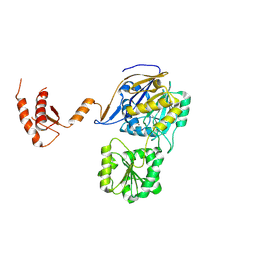 | | RNase J from Mycobacterium tuberculosis | | 分子名称: | Ribonuclease J, ZINC ION | | 著者 | Li, J, Bao, L, Hu, J, Zhan, B, Li, Z. | | 登録日 | 2022-01-19 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
7WNU
 
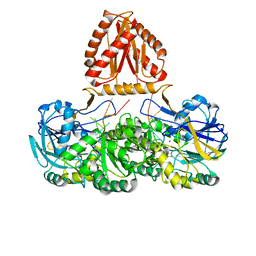 | | Mycobacterium tuberculosis Rnase J complex with 7nt RNA | | 分子名称: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease J, ZINC ION | | 著者 | Li, J, Hu, J, Bao, L, Zhan, B, Li, Z. | | 登録日 | 2022-01-19 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-09-04 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
2FCO
 
 | | Crystal Structure of Bacillus stearothermophilus PrfA-Holliday Junction Resolvase | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, recombination protein U (penicillin-binding protein related factor A) | | 著者 | Li, J, Jedrzejas, M.J. | | 登録日 | 2005-12-12 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure, flexibility, and mechanism of the Bacillus stearothermophilus RecU Holliday junction resolvase.
Proteins, 68, 2007
|
|
8U9J
 
 | | The crystal structure of iron-bound human ADO C18S C239S variant at 2.02 Angstrom | | 分子名称: | 2-aminoethanethiol dioxygenase, FE (III) ION, GLYCEROL, ... | | 著者 | Liu, A, Li, J, Duan, R. | | 登録日 | 2023-09-19 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Cobalt(II)-Substituted Cysteamine Dioxygenase Oxygenation Proceeds through a Cobalt(III)-Superoxo Complex.
J.Am.Chem.Soc., 146, 2024
|
|
8UAN
 
 | | The crystal structure of cobalt-bound human ADO C18S C239S variant at 1.99 Angstrom | | 分子名称: | 2-aminoethanethiol dioxygenase, COBALT (II) ION, GLYCEROL | | 著者 | Liu, A, Li, J, Duan, R, Shin, I. | | 登録日 | 2023-09-21 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Cobalt(II)-Substituted Cysteamine Dioxygenase Oxygenation Proceeds through a Cobalt(III)-Superoxo Complex.
J.Am.Chem.Soc., 146, 2024
|
|
4MLO
 
 | | 1.65A resolution structure of ToxT from Vibrio cholerae (P21 Form) | | 分子名称: | CHLORIDE ION, PALMITOLEIC ACID, TCP pilus virulence regulatory protein | | 著者 | Lovell, S, Wehmeyer, G, Battaile, K.P, Li, J, Egan, S. | | 登録日 | 2013-09-06 | | 公開日 | 2016-04-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | 1.65 angstrom resolution structure of the AraC-family transcriptional activator ToxT from Vibrio cholerae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
6BPR
 
 | | Crystal structure of cysteine, nitric oxide-bound ferrous form of the uncrosslinked F2-Tyr157 human cysteine dioxygenase | | 分子名称: | CYSTEINE, Cysteine dioxygenase type 1, FE (III) ION, ... | | 著者 | Liu, A, Li, J, Shin, I. | | 登録日 | 2017-11-26 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Probing the Cys-Tyr Cofactor Biogenesis in Cysteine Dioxygenase by the Genetic Incorporation of Fluorotyrosine.
Biochemistry, 58, 2019
|
|
6DKD
 
 | | Yeast Ddi2 Cyanamide Hydratase | | 分子名称: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | 著者 | Moore, S.A, Xiao, W, Li, J. | | 登録日 | 2018-05-29 | | 公開日 | 2019-05-08 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
5DTH
 
 | |
4PY4
 
 | |
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | 分子名称: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
7CFU
 
 | |
2L26
 
 | | Rv0899 from Mycobacterium tuberculosis contains two separated domains | | 分子名称: | Uncharacterized protein Rv0899/MT0922 | | 著者 | Shi, C, Li, J, Gao, Y, Wu, K, Huang, H, Tian, C. | | 登録日 | 2010-08-12 | | 公開日 | 2011-08-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Studies of Mycobacterium tuberculosis Rv0899 Reveal a Monomeric Membrane-Anchoring Protein with Two Separate Domains
J.Mol.Biol., 2011
|
|
7VVP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | 登録日 | 2021-11-07 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7WQJ
 
 | | Crystal structure of MERS main protease in complex with PF07304814 | | 分子名称: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | 著者 | Lin, C, Zhang, J, Li, J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
7CDB
 
 | | Structure of GABARAPL1 in complex with GABA(A) receptor gamma 2 | | 分子名称: | ACETATE ION, CITRIC ACID, Gamma-aminobutyric acid receptor subunit gamma-2, ... | | 著者 | Li, J, Ye, J, Zhu, R, Kong, C, Zhang, M, Wang, C. | | 登録日 | 2020-06-19 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Structural basis of GABARAP-mediated GABA A receptor trafficking and functions on GABAergic synaptic transmission.
Nat Commun, 12, 2021
|
|
7VLQ
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.939106 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.0227 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | 登録日 | 2021-10-05 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.50251937 Å) | | 主引用文献 | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
