7WAX
 
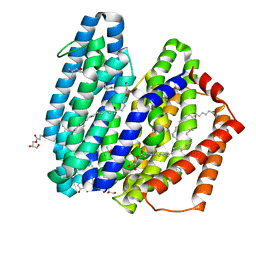 | | MurJ inward occluded form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, lipid II flippase MurJ | | 著者 | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | 登録日 | 2021-12-15 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAG
 
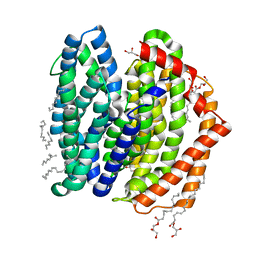 | | Crystal structure of MurJ squeezed form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Lipid II flippase MurJ | | 著者 | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | 登録日 | 2021-12-14 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
7WAW
 
 | | MurJ inward closed form | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, lipid II flippase MurJ | | 著者 | Tsukazaki, T, Kohga, H, Tanaka, Y, Yoshikaie, K, Taniguchi, K, Fujimoto, K. | | 登録日 | 2021-12-15 | | 公開日 | 2022-06-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the lipid flippase MurJ in a "squeezed" form distinct from its inward- and outward-facing forms.
Structure, 30, 2022
|
|
4OZQ
 
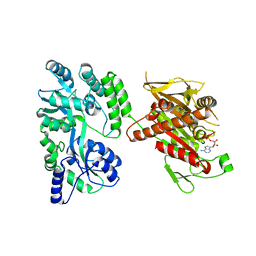 | | Crystal structure of the mouse Kif14 motor domain | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Chimera of Maltose-binding periplasmic protein and Kinesin family member 14 protein | | 著者 | Arora, K, Talje, L, Asenjo, A.B, Andersen, P, Atchia, K, Joshi, M, Sosa, H, Kwok, B.H, Allingham, J.S. | | 登録日 | 2014-02-18 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | KIF14 binds tightly to microtubules and adopts a rigor-like conformation.
J.Mol.Biol., 426, 2014
|
|
1IQW
 
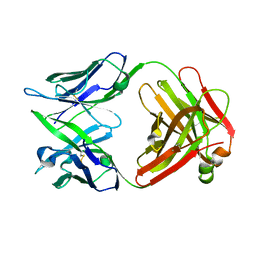 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A | | 分子名称: | ANTIBODY M-HFE7A, HEAVY CHAIN, LIGHT CHAIN | | 著者 | Ito, S, Takayama, T, Hanzawa, H, Ichikawa, K, Ohsumi, J, Serizawa, N, Hata, T, Haruyama, H. | | 登録日 | 2001-08-10 | | 公開日 | 2002-01-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the antigen-binding fragment of apoptosis-inducing mouse anti-human Fas monoclonal antibody HFE7A.
J.Biochem., 131, 2002
|
|
1PRU
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | 分子名称: | PURINE REPRESSOR | | 著者 | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | 登録日 | 1995-05-08 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
1PRV
 
 | | PURINE REPRESSOR DNA-BINDING DOMAIN DNA BINDING | | 分子名称: | PURINE REPRESSOR | | 著者 | Nagadoi, A, Morikawa, S, Nakamura, H, Enari, M, Kobayashi, K, Yamamoto, H, Sampei, G, Mizobuchi, K, Schumacher, M.A, Brennan, R.G, Nishimura, Y. | | 登録日 | 1995-05-08 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural comparison of the free and DNA-bound forms of the purine repressor DNA-binding domain.
Structure, 3, 1995
|
|
8DJI
 
 | | Ternary complex of SUMO1 with the SIM of PML and zinc | | 分子名称: | Protein PML, Small ubiquitin-related modifier 1, ZINC ION | | 著者 | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | 登録日 | 2022-06-30 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
6L98
 
 | | Crystalline cast nephropathy-causing Bence-Jones protein AK: An entire immunoglobulin lambda light chain dimer | | 分子名称: | Bence-Jones protein lambda light chain AK | | 著者 | Nakagaki, T, Noguchi, K, Yohda, M, Odaka, M, Wakui, H, Matsumura, H. | | 登録日 | 2019-11-08 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Multiple Myeloma-Associated Ig Light Chain Crystalline Cast Nephropathy.
Kidney Int Rep, 5, 2020
|
|
5YGX
 
 | | Structure of BACE1 in complex with N-(3-((4R,5R,6S)-2-amino-6-(1,1-difluoroethyl)-5-fluoro-4-methyl-5,6-dihydro-4H-1,3-oxazin-4-yl)-4-fluorophenyl)-5-(fluoromethoxy)pyrazine-2-carboxamide | | 分子名称: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Nakahara, K, Fuchino, K, Komano, K, Asada, N, Tadano, G, Hasegawa, T, Yamamoto, T, Sako, Y, Ogawa, M, Unemura, C, Hosono, M, Sakaguchi, G, Ando, S, Ohnishi, S, Kido, Y, Fukushima, T, Dhuyvetter, D, Borghys, H, Gijsen, H, Yamano, Y, Iso, Y, Kusakabe, K. | | 登録日 | 2017-09-27 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of Potent and Centrally Active 6-Substituted 5-Fluoro-1,3-dihydro-oxazine beta-Secretase (BACE1) Inhibitors via Active Conformation Stabilization
J. Med. Chem., 61, 2018
|
|
8DJH
 
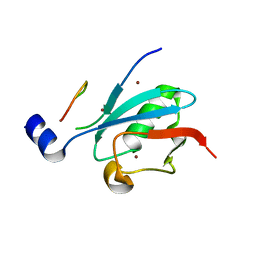 | | Ternary complex of SUMO1 with a phosphomimetic SIM of PML and zinc | | 分子名称: | PML 4SD, Small ubiquitin-related modifier 1, ZINC ION | | 著者 | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Bourdeau, V, Gagnon, C, Igelmann, S, Sakaguchi, K, Ferbeyre, G, Omichinski, J.G. | | 登録日 | 2022-06-30 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Zinc controls PML nuclear body formation through regulation of a paralog specific auto-inhibition in SUMO1.
Nucleic Acids Res., 50, 2022
|
|
1EQ5
 
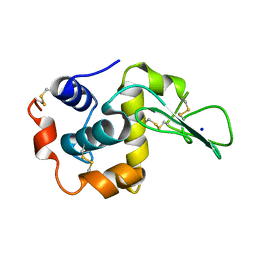 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
1EQE
 
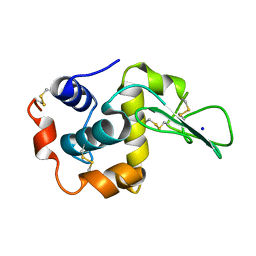 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-04-04 | | 公開日 | 2000-04-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
2D3H
 
 | | Crystal structures of collagen model peptides (Pro-Pro-Gly)4-Hyp-Hyp-Gly-(Pro-Pro-Gly)4 | | 分子名称: | COLLAGEN MODEL PEPTIDES (PRO-PRO-GLY)4-HYP-HYP-GLY-(PRO-PRO-GLY)4 | | 著者 | Wu, G, Noguchi, K, Okuyama, K, Mizuno, K, Bachinger, H.P. | | 登録日 | 2005-09-28 | | 公開日 | 2006-09-19 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | High-resolution structures of collagen-like peptides [(Pro-Pro-Gly)(4)-Xaa-Yaa-Gly-(Pro-Pro-Gly)(4)]: Implications for triple-helix hydration and Hyp(X) puckering.
Biopolymers, 91, 2009
|
|
8COG
 
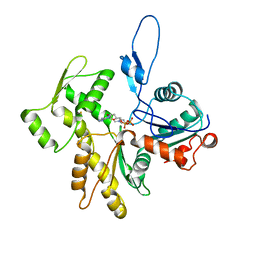 | | Human arginylated beta-actin | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | 著者 | Pinto, C.S, Bakker, S.E, Suchenko, A, Hussain, H, Hatano, T, Sampath, K, Chinthalapudi, K, Mishima, M, Balasubramanian, M. | | 登録日 | 2023-02-28 | | 公開日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.499 Å) | | 主引用文献 | Structure and physiological investigation of human arginylated beta-actin
To Be Published
|
|
1UAT
 
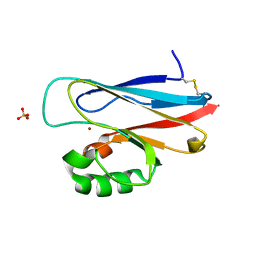 | | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J | | 分子名称: | Azurin iso-2, COPPER (II) ION, SULFATE ION | | 著者 | Inoue, T, Suzuki, S, Nisho, N, Yamaguchi, K, Kataoka, K, Tobari, J, Yong, X, Hamanaka, S, Matsumura, H, Kai, Y. | | 登録日 | 2003-03-19 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J
J.Mol.Biol., 333, 2003
|
|
6U4K
 
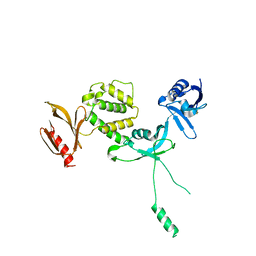 | | Human talin2 residues 1-403 | | 分子名称: | Talin-2 | | 著者 | Izard, T, Rangarajan, E.S, Colgan, L, Yasuda, R, Chinthalapudi, K. | | 登録日 | 2019-08-26 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-23 | | 実験手法 | X-RAY DIFFRACTION (2.555 Å) | | 主引用文献 | A distinct talin2 structure directs isoform specificity in cell adhesion.
J.Biol.Chem., 295, 2020
|
|
2AEN
 
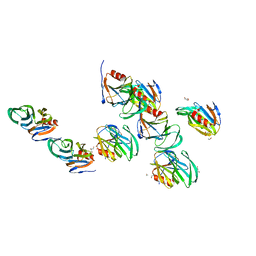 | | Crystal structure of the rotavirus strain DS-1 VP8* core | | 分子名称: | ETHANOL, GLYCEROL, Outer capsid protein VP4, ... | | 著者 | Monnier, N, Higo-Moriguchi, K, Sun, Z.-Y.J, Prasad, B.V.V, Taniguchi, K, Dormitzer, P.R. | | 登録日 | 2005-07-22 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.604 Å) | | 主引用文献 | High-resolution molecular and antigen structure of the VP8*
core of a sialic acid-independent human rotavirus strain
J.Virol., 80, 2006
|
|
1EQ4
 
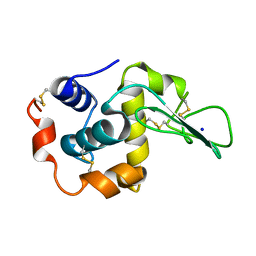 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | 分子名称: | LYSOZYME, SODIUM ION | | 著者 | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-19 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
3ED9
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [30 min] | | 分子名称: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | 登録日 | 2008-09-02 | | 公開日 | 2009-02-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1GA6
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSCP (PSEUDOMONAS SERINE-CARBOXYL PROTEINASE) COMPLEXED WITH A FRAGMENT OF TYROSTATIN (THIS ENZYME RENAMED "SEDOLISIN" IN 2003) | | 分子名称: | ACETATE ION, CALCIUM ION, FRAGMENT OF TYROSTATIN, ... | | 著者 | Wlodawer, A, Li, M, Dauter, Z, Gustchina, A, Uchida, K. | | 登録日 | 2000-11-29 | | 公開日 | 2000-12-13 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes.
Nat.Struct.Biol., 8, 2001
|
|
6EN6
 
 | | Crystal structure B of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | 分子名称: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | 登録日 | 2017-10-04 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
2E1Q
 
 | | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | | 分子名称: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | 著者 | Yamaguchi, Y, Matsumura, T, Ichida, K, Okamoto, K, Nishino, T. | | 登録日 | 2006-10-27 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate
J.Biochem.(Tokyo), 141, 2007
|
|
6EN5
 
 | | Crystal structure A of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | 分子名称: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | 登録日 | 2017-10-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
7CG3
 
 | | Staggered ring conformation of CtHsp104 (Hsp104 from Chaetomium Thermophilum) | | 分子名称: | Heat shock protein 104 | | 著者 | Inoue, Y, Hanazono, Y, Noi, K, Kawamoto, A, Kimatsuka, M, Harada, R, Takeda, K, Iwamasa, N, Shibata, K, Noguchi, K, Shigeta, Y, Namba, K, Ogura, T, Miki, K, Shinohara, K, Yohda, M. | | 登録日 | 2020-06-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Split conformation of Chaetomium thermophilum Hsp104 disaggregase.
Structure, 29, 2021
|
|
