6WIP
 
 | | Class A beta-lactamase from Micromonospora aurantiaca ATCC 27029 | | 分子名称: | ACETATE ION, Beta-lactamase, SULFATE ION | | 著者 | Chang, C, Tesar, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-22 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | class A beta-lactamase from
Micromonospora aurantiaca ATCC 27029
To Be Published
|
|
5FDA
 
 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | 分子名称: | CHLORIDE ION, Dihydrofolate reductase | | 著者 | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-12-15 | | 公開日 | 2015-12-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
5EQC
 
 | | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | 著者 | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-12 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site
To Be Published
|
|
5HJ1
 
 | | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid | | 分子名称: | Pullulanase C protein, VACCENIC ACID | | 著者 | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-01-12 | | 公開日 | 2016-02-17 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid
To Be Published
|
|
5HQH
 
 | | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes. | | 分子名称: | CHLORIDE ION, Lmo2119 protein, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-01-21 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes.
To Be Published
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | 分子名称: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | 著者 | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-01-07 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
5HZL
 
 | |
6UX3
 
 | | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae | | 分子名称: | Acetoin dehydrogenase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | 著者 | Chang, C, Skarina, T, Mesa, N, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-11-06 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Crystal structure of acetoin dehydrogenase from Enterobacter cloacae
To Be Published
|
|
6VBF
 
 | | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii | | 分子名称: | CALCIUM ION, CHLORIDE ION, Two-component regulatory system response regulator | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-18 | | 公開日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | 1.85 Angstrom Resolution Crystal Structure of N-terminal Domain of Two-component System Response Regulator from Acinetobacter baumannii
To Be Published
|
|
6VJ2
 
 | | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis | | 分子名称: | Foldase | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-01-14 | | 公開日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | 3.10 Angstrom Resolution Crystal Structure of Foldase Protein (PrsA) from Lactococcus lactis
To Be Published
|
|
6VJ4
 
 | | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis | | 分子名称: | Peptidylprolyl isomerase PrsA | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-01-14 | | 公開日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | 1.70 Angstrom Resolution Crystal Structure of Peptidylprolyl Isomerase (PrsA) from Bacillus anthracis
To Be Published
|
|
6VPU
 
 | | 1.90 Angstrom Resolution Crystal Structure Phosphoadenosine Phosphosulfate Reductase (CysH) from Vibrio vulnificus | | 分子名称: | DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, Phosphoadenosine phosphosulfate reductase, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-04 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 1.90 Angstrom Resolution Crystal Structure Phosphoadenosine Phosphosulfate Reductase (CysH) from Vibrio vulnificus
To Be Published
|
|
6WJM
 
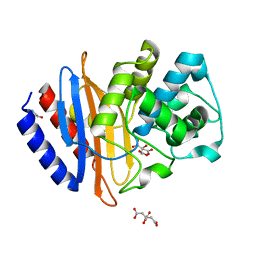 | | The crystal structure beta-lactamase from Desulfarculus baarsii DSM 2075 | | 分子名称: | ALANINE, Beta-lactamase, CITRIC ACID | | 著者 | Chang, C, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-14 | | 公開日 | 2020-04-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | The crystal structure beta-lactamase from Desulfarculus baarsii DSM 2075
To Be Published
|
|
6X4I
 
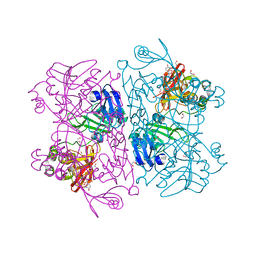 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | 分子名称: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | 著者 | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-22 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WY4
 
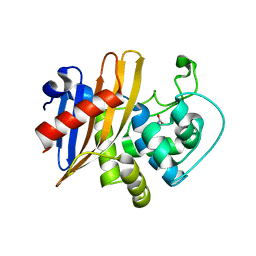 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-12 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
6WXC
 
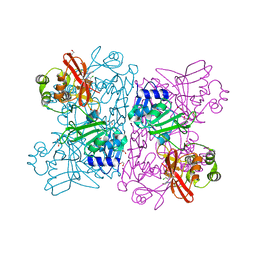 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with potential repurposing drug Tipiracil | | 分子名称: | 1,2-ETHANEDIOL, 5-CHLORO-6-(1-(2-IMINOPYRROLIDINYL) METHYL) URACIL, FORMIC ACID, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-10 | | 公開日 | 2020-05-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WLC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-19 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6X1B
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-18 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6XD8
 
 | | Crystal Structure of Peptidylprolyl Isomerase (PrsA) Fragment from Bacillus anthracis | | 分子名称: | Foldase protein PrsA 1 | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-10 | | 公開日 | 2020-07-01 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal Structure of Peptidylprolyl Isomerase (PrsA) Fragment from Bacillus anthracis
To Be Published
|
|
5HL8
 
 | | 1.93 Angstrom resolution crystal structure of a pullulanase-specific type II secretion system integral cytoplasmic membrane protein GspL (C-terminal fragment; residues 309-397) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | 分子名称: | Type II secretion system protein L | | 著者 | Halavaty, A.S, Minasov, G, Kiryukhina, O, Grimshaw, S, Light, S, Dubrovska, I, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-01-14 | | 公開日 | 2016-01-27 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | 1.93 Angstrom resolution crystal structure of a pullulanase-specific type II secretion system integral cytoplasmic membrane protein GspL (C-terminal fragment; residues 309-397) from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044
To Be Published
|
|
6VPW
 
 | | 1.90 Angstrom Resolution Crystal Structure Chemotaxis protein CheX from Vibrio vulnificus | | 分子名称: | Chemotaxis protein CheX | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Wiersum, G, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-02-04 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 1.90 Angstrom Resolution Crystal Structure Chemotaxis protein CheX from Vibrio vulnificus
To Be Published
|
|
7MTX
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P176 | | 分子名称: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-beta-D-ribopyranosylamine, ... | | 著者 | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-05-13 | | 公開日 | 2021-06-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P176
To Be Published
|
|
3OUT
 
 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | 分子名称: | D-GLUTAMIC ACID, Glutamate racemase | | 著者 | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-15 | | 公開日 | 2010-09-29 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
3OF5
 
 | | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4 | | 分子名称: | ACETATE ION, Dethiobiotin synthetase, SODIUM ION | | 著者 | Brunzelle, J.S, Skarina, T, Gordon, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-08-13 | | 公開日 | 2011-02-02 | | 最終更新日 | 2018-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal Structure of a Dethiobiotin Synthetase from Francisella tularensis subsp. tularensis SCHU S4
TO BE PUBLISHED
|
|
6B4O
 
 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | 著者 | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-09-27 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
