5BW0
 
 | | The crystal structure of minor pseudopilin binary complex of XcpV and XcpW from the Type 2 secretion system of Pseudomonas aeruginosa | | Descriptor: | SULFATE ION, Type II secretion system protein I, Type II secretion system protein J | | Authors: | Zhang, Y, Faucher, F, Poole, K, Jia, Z. | | Deposit date: | 2015-06-05 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided disruption of the pseudopilus tip complex inhibits the Type II secretion in Pseudomonas aeruginosa.
PLoS Pathog., 14, 2018
|
|
2P6W
 
 | | Crystal structure of a glycosyltransferase involved in the glycosylation of the major capsid of PBCV-1 | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, Putative glycosyltransferase (Mannosyltransferase) involved in glycosylating the PBCV-1 major capsid protein | | Authors: | Zhang, Y, Ye, X, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2007-03-19 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and function of a chlorella virus-encoded glycosyltransferase.
Structure, 15, 2007
|
|
1TZ3
 
 | | crystal structure of aminoimidazole riboside kinase complexed with aminoimidazole riboside | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOSIDE, POTASSIUM ION, putative sugar kinase | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
1U7D
 
 | |
1TYY
 
 | | Crystal structure of aminoimidazole riboside kinase from Salmonella enterica | | Descriptor: | POTASSIUM ION, putative sugar kinase | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-08 | | Release date: | 2004-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
1TZ6
 
 | | Crystal structure of aminoimidazole riboside kinase from Salmonella enterica complexed with aminoimidazole riboside and ATP analog | | Descriptor: | 5-AMINOIMIDAZOLE RIBONUCLEOSIDE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Zhang, Y, Dougherty, M, Downs, D.M, Ealick, S.E. | | Deposit date: | 2004-07-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of an Aminoimidazole Riboside Kinase from Salmonella enterica; Implications for the Evolution of the Ribokinase Superfamily
STRUCTURE, 12, 2004
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
1FBC
 
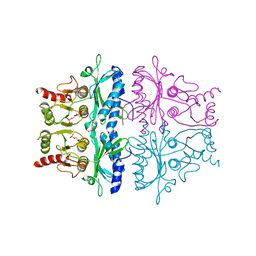 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBG
 
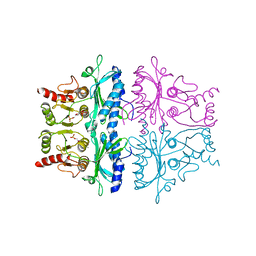 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBF
 
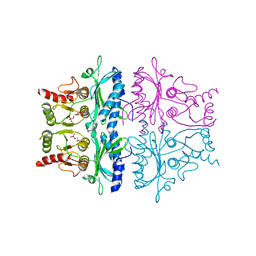 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1FBD
 
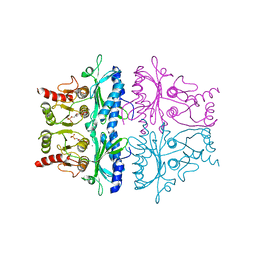 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1THD
 
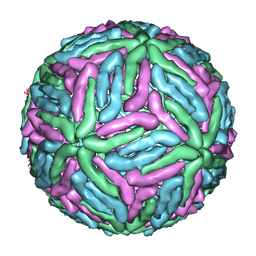 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
5E00
 
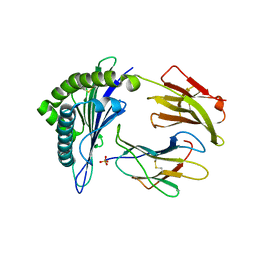 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, GLY-VAL-TRP-ILE-ARG-THR-PRO-PRO-ALA, HLA class I histocompatibility antigen, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2015-09-26 | | Release date: | 2017-01-18 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
3OMX
 
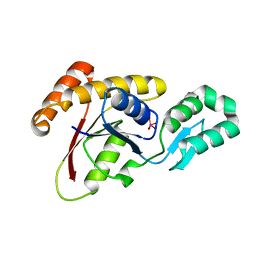 | | Crystal structure of Ssu72 with vanadate complex | | Descriptor: | CG14216, VANADATE ION | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3366 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
3OMW
 
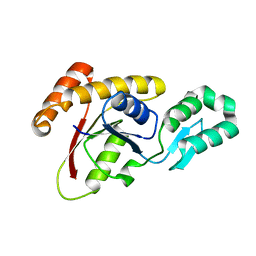 | | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II | | Descriptor: | CG14216 | | Authors: | Zhang, Y, Zhang, M, Zhang, Y. | | Deposit date: | 2010-08-27 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8701 Å) | | Cite: | Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue.
Biochem.J., 434, 2011
|
|
1N6G
 
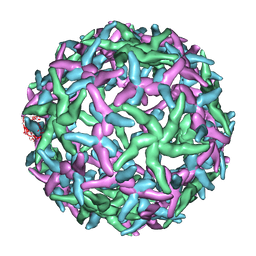 | | The structure of immature Dengue-2 prM particles | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
1NA4
 
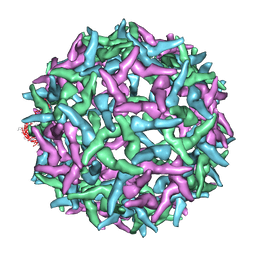 | | The structure of immature Yellow Fever virus particle | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
1VZX
 
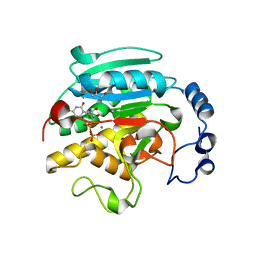 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-28 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
1FNL
 
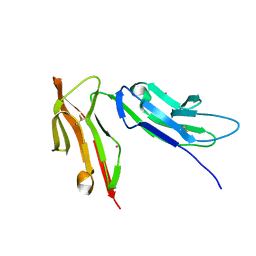 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF A HUMAN FCGRIII | | Descriptor: | LOW AFFINITY IMMUNOGLOBULIN GAMMA FC REGION RECEPTOR III-B, MERCURY (II) ION | | Authors: | Zhang, Y, Boesen, C.C, Radaev, S, Brooks, A.G, Fridman, W.H, Sautes-Fridman, C, Sun, P.D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the extracellular domain of a human Fc gamma RIII.
Immunity, 13, 2000
|
|
1VZU
 
 | | Roles of active site tryptophans in substrate binding and catalysis by ALPHA-1,3 GALACTOSYLTRANSFERASE | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Deshpande, A, Xie, Z, Natesh, R, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-27 | | Release date: | 2004-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of active site tryptophans in substrate binding and catalysis by alpha-1,3 galactosyltransferase.
Glycobiology, 14, 2004
|
|
1FKC
 
 | | HUMAN PRION PROTEIN (MUTANT E200K) FRAGMENT 90-231 | | Descriptor: | PRION PROTEIN | | Authors: | Zhang, Y, Swietnicki, W, Zagorski, M.G, Surewicz, W.K, Soennichsen, F.D. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-21 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the E200K variant of human prion protein. Implications for the mechanism of pathogenesis in familial prion diseases.
J.Biol.Chem., 275, 2000
|
|
6O7G
 
 | |
4KMH
 
 | |
4KM8
 
 | | Crystal structure of Sufud60 | | Descriptor: | Suppressor of fused homolog | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
4KMA
 
 | |
