4U4P
 
 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
2AI5
 
 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-07-29 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
1WZB
 
 | | Crystal structure of the collagen triple helix model [{HYP(R)-HYP(R)-GLY}10]3 | | Descriptor: | Collagen triple helix | | Authors: | Kawahara, K, Nakamura, S, Nishi, Y, Uchiyama, S, Nishiuchi, Y, Nakazawa, T, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effect of hydration on the stability of the collagen-like triple-helical structure of [4(R)-hydroxyprolyl-4(R)-hydroxyprolylglycine]10
Biochemistry, 44, 2005
|
|
7BPG
 
 | | Structure of serinol nucleic acid - RNA complex | | Descriptor: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7BPF
 
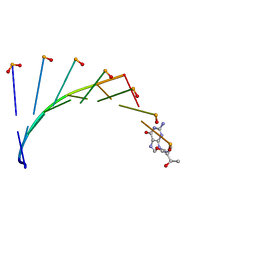 | | Structure of L-threoninol nucleic acid - RNA complex | | Descriptor: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
1ISE
 
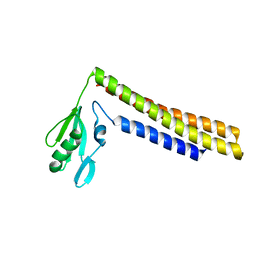 | | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly | | Descriptor: | Ribosome Recycling Factor | | Authors: | Nakano, H, Yoshida, T, Oka, S, Uchiyama, S, Nishina, K, Ohkubo, T, Kato, H, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly
To be Published
|
|
1IS1
 
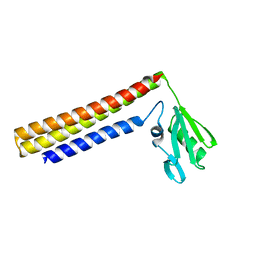 | | Crystal structure of ribosome recycling factor from Vibrio parahaemolyticus | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Nakano, H, Yamaichi, Y, Uchiyama, S, Yoshida, T, Nishina, K, Kato, H, Ohkubo, T, Honda, T, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium
J.BIOL.CHEM., 278, 2003
|
|
2DT4
 
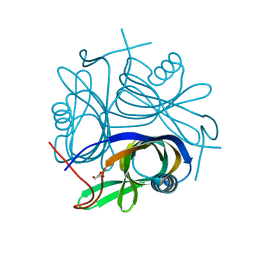 | | Crystal structure of Pyrococcus horikoshii a plant- and prokaryote-conserved (PPC) protein at 1.60 resolution | | Descriptor: | GLYCEROL, Hypothetical protein PH0802 | | Authors: | Lin, L, Nakano, H, Uchiyama, S, Fujimoto, S, Matsunaga, S, Nakamura, S. | | Deposit date: | 2006-07-10 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Pyrococcus horikoshii PPC protein at 1.60 A resolution
Proteins, 67, 2007
|
|
7W64
 
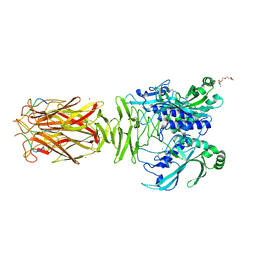 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with N-terminal peptide fragment of TcpF | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W65
 
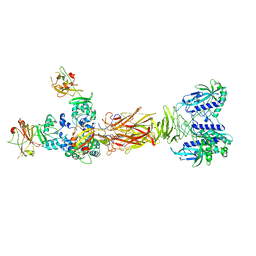 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with secreted protein TcpF | | Descriptor: | Toxin coregulated pilus biosynthesis protein F, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W63
 
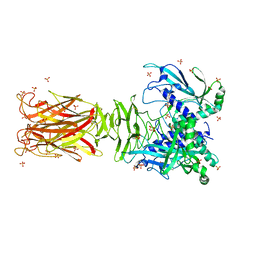 | | Crystal structure of minor pilin TcpB from Vibrio cholerae | | Descriptor: | SULFATE ION, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
2D0S
 
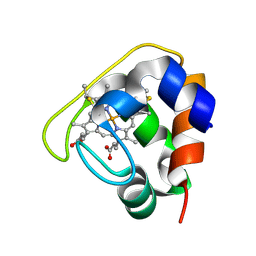 | | Crystal structure of the Cytochrome C552 from moderate thermophilic bacterium, hydrogenophilus thermoluteolus | | Descriptor: | HEME C, cytochrome c | | Authors: | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | Deposit date: | 2005-08-08 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
1DVV
 
 | | SOLUTION STRUCTURE OF THE QUINTUPLE MUTANT OF CYTOCHROME C-551 FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hasegawa, J, Uchiyama, S, Tanimoto, Y, Mizutani, M, Kobayashi, Y, Sambongi, Y, Igarashi, Y. | | Deposit date: | 2000-01-22 | | Release date: | 2000-11-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Selected mutations in a mesophilic cytochrome c confer the stability of a thermophilic counterpart.
J.Biol.Chem., 275, 2000
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | Deposit date: | 2000-10-19 | | Release date: | 2001-05-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
5JQP
 
 | | Crystal structure of ER glucosidase II heterodimeric complex consisting of catalytic subunit and the binding domain of regulatory subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CALCIUM ION, ... | | Authors: | Satoh, T, Toshimori, T, Noda, M, Uchiyama, S, Kato, K. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction mode between catalytic and regulatory subunits in glucosidase II involved in ER glycoprotein quality control.
Protein Sci., 25, 2016
|
|
5ZRS
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl adipate bound state | | Descriptor: | 6-ethoxy-6-oxohexanoic acid, Alpha/beta hydrolase family protein, CALCIUM ION, ... | | Authors: | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRR
 
 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in monoethyl succinate bound state | | Descriptor: | 4-ethoxy-4-oxobutanoic acid, Alpha/beta hydrolase family protein, GLYCEROL, ... | | Authors: | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZRQ
 
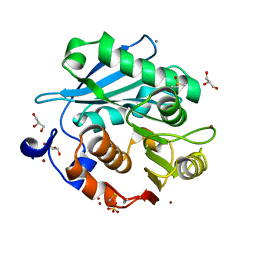 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S mutant in Zn(2+)-bound state | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL, ... | | Authors: | Numoto, N, Kamiya, N, Bekker, G.J, Yamagami, Y, Inaba, S, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
5ZNO
 
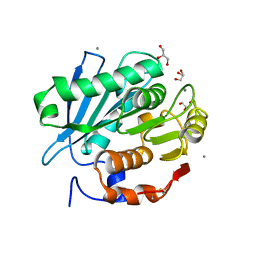 | | Crystal structure of PET-degrading cutinase Cut190 S176A/S226P/R228S/ mutant in Ca(2+)-bound state | | Descriptor: | Alpha/beta hydrolase family protein, CALCIUM ION, GLYCEROL | | Authors: | Numoto, N, Inaba, S, Yamagami, Y, Kamiya, N, Bekker, G.J, Ishii, K, Uchiyama, S, Kawai, F, Ito, N, Oda, M. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.60264349 Å) | | Cite: | Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle.
Biochemistry, 57, 2018
|
|
3AHA
 
 | | Crystal structure of the complex between gp41 fragments N36 and C34 mutant N126K/E137Q | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Transmembrane protein gp41 | | Authors: | Izumi, K, Nakamura, S, Nakano, H, Shimura, K, Sakagami, Y, Oishi, S, Uchiyama, S, Ohkubo, T, Kobayashi, Y, Fujii, N, Matsuoka, M, Kodama, E.N. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino terminal heptad repeat.
Antiviral Res., 2010
|
|
3A4U
 
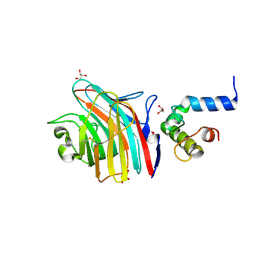 | | Crystal structure of MCFD2 in complex with carbohydrate recognition domain of ERGIC-53 | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Nishio, M, Kamiya, Y, Mizushima, T, Wakatsuki, S, Sasakawa, H, Yamamoto, K, Uchiyama, S, Noda, M, McKay, A.R, Fukui, K, Hauri, H.P, Kato, K. | | Deposit date: | 2009-07-17 | | Release date: | 2010-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | Descriptor: | Collagen-like peptide | | Authors: | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | Deposit date: | 2011-07-26 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
3WJ4
 
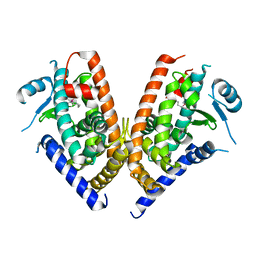 | | Crystal structure of PPARgamma ligand binding domain in complex with tributyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, tributylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
3WJ5
 
 | | Crystal structure of PPARgamma ligand binding domain in complex with triphenyltin | | Descriptor: | Peroxisome proliferator-activated receptor gamma, triphenylstannanyl | | Authors: | Harada, S, Hiromori, Y, Fukakusa, S, Kawahara, K, Nakamura, S, Noda, M, Uchiyama, S, Fukui, K, Nishikawa, J, Nagase, H, Kobayashi, Y, Ohkubo, T, Yoshida, T, Nakanishi, T. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for PPARgamma transactivation by endocrine disrupting organotin compounds
To be Published
|
|
