5HDM
 
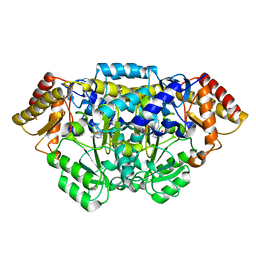 | | Crystal structure of Arabidopsis thaliana glutamate-1-semialdehyde-2,1-aminomutase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase 1, chloroplastic, ... | | Authors: | Song, Y, Pu, H, Liu, L. | | Deposit date: | 2016-01-05 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of glutamate-1-semialdehyde-2,1-aminomutase from Arabidopsis thaliana.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4TTU
 
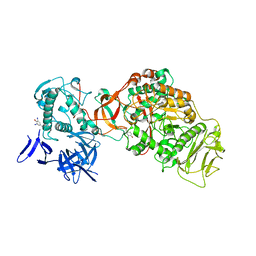 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with isomaltotriose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Dextransucrase, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
4TVC
 
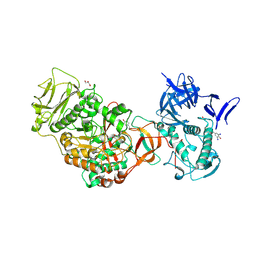 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with gluco-oligosaccharides | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
4TVD
 
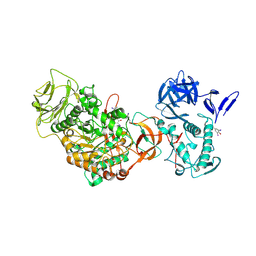 | | N-terminally truncated dextransucrase DSR-E from Leuconostoc mesenteroides NRRL B-1299 in complex with D-glucose | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Dextransucrase, ... | | Authors: | Brison, Y, Remaud-Simeon, M, Mourey, L, Tranier, S. | | Deposit date: | 2014-06-26 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Carbohydrate Binding Ability of an alpha-(12) Branching Sucrase from Glycoside Hydrolase Family 70.
J.Biol.Chem., 291, 2016
|
|
6J3A
 
 | | Structure of LmbA2991HD | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Song, Y. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of LmbA2991HD
To Be Published
|
|
6J3I
 
 | | Structure of LmbA2991T421A | | Descriptor: | Gamma-glutamyltranspeptidase | | Authors: | Song, Y. | | Deposit date: | 2019-01-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of LmbA2991T421A
To Be Published
|
|
2YFY
 
 | | SERCA in the HnE2 State Complexed With Debutanoyl Thapsigargin | | Descriptor: | DEBUTANOYL THAPSIGARGIN, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sonntag, Y, Musgaard, M, Olesen, C, Schiott, B, Moller, J.V, Nissen, P, Thogersen, L. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mutual Adaptation of a Membrane Protein and its Lipid Bilayer During Conformational Changes.
Nat.Commun., 2, 2011
|
|
5X7L
 
 | |
5YH1
 
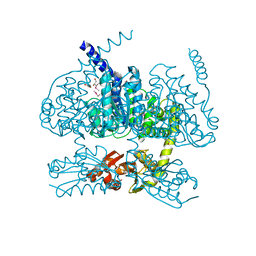 | |
5CL2
 
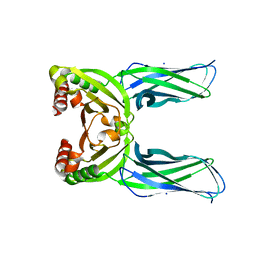 | | Crystal structure of Spo0M, sporulation control protein, from Bacillus subtilis. | | Descriptor: | SODIUM ION, Sporulation-control protein spo0M | | Authors: | Sonoda, Y, Mizutani, K, Mikami, B. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Spo0M, a sporulation-control protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3Q72
 
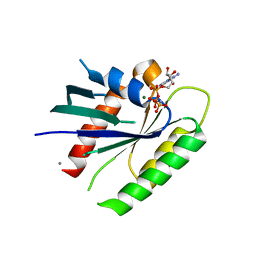 | | Crystal Structure of Rad G-domain-GTP Analog Complex | | Descriptor: | CALCIUM ION, GTP-binding protein RAD, MAGNESIUM ION, ... | | Authors: | Sasson, Y, Navon-Perry, L, Hirsch, J.A. | | Deposit date: | 2011-01-04 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | RGK Family G-Domain:GTP Analog Complex Structures and Nucleotide-Binding Properties.
J.Mol.Biol., 413, 2011
|
|
3Q7P
 
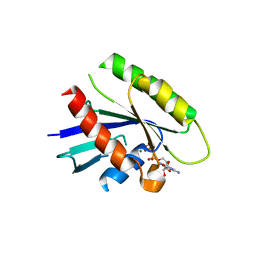 | |
3TTQ
 
 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
3TTO
 
 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in triclinic form | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Tranier, S, Remaud-Simeon, M, Dijkstra, B.W. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
3Q7Q
 
 | | Crystal Structure of Rad G-domain Q148A-GTP Analog Complex | | Descriptor: | CALCIUM ION, GTP-binding protein RAD, MAGNESIUM ION, ... | | Authors: | Sasson, Y, Navon-Perry, L, Hirsch, J.A. | | Deposit date: | 2011-01-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RGK Family G-Domain:GTP Analog Complex Structures and Nucleotide-Binding Properties.
J.Mol.Biol., 413, 2011
|
|
3SOE
 
 | | Crystal Structure of the 3rd PDZ domain of the human Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3 (MAGI3) | | Descriptor: | 1,2-ETHANEDIOL, Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3 | | Authors: | Ivarsson, Y, Filippakopoulos, P, Picaud, S, Vollmar, M, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S, Zimmermann, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the 3rd PDZ domain of the human Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3 (MAGI3)
To be Published
|
|
1R62
 
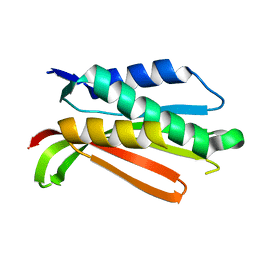 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | Descriptor: | Nitrogen regulation protein NR(II) | | Authors: | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | Deposit date: | 2003-10-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
8UBZ
 
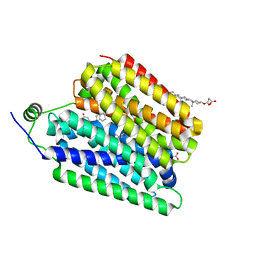 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBX
 
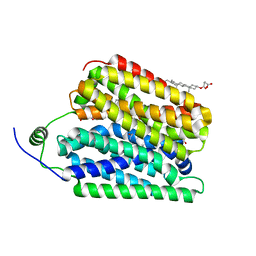 | | Ethanolamine-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, ETHANOLAMINE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBY
 
 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UC0
 
 | | Endogenous ligand bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
8UBW
 
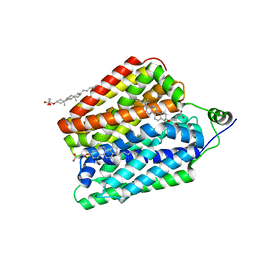 | | Choline-bound FLVCR1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, CHOLINE ION, Heme transporter FLVCR1 | | Authors: | Hite, R.K, Son, Y. | | Deposit date: | 2023-09-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural basis of lipid head group entry to the Kennedy pathway by FLVCR1.
Nature, 629, 2024
|
|
7V1Q
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
7V1R
 
 | | Leifsonia Alcohol Dehydrogenases LnADH | | Descriptor: | Alcohol Dehydrogenases, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Song, Y, Qu, X. | | Deposit date: | 2021-08-05 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Engineering Leifsonia Alcohol Dehydrogenase for Thermostability and Catalytic Efficiency by Enhancing Subunit Interactions.
Chembiochem, 22, 2021
|
|
6ITX
 
 | |
