9DUB
 
 | | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound SRM-25-071 | | Descriptor: | 4-(pyridin-4-yl)pyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Minasov, G, Winsor, J, Roy, S.M, Watterson, D.M, Shuvalova, L. | | Deposit date: | 2024-10-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound SRM-25-071
To Be Published
|
|
5KZ6
 
 | | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Chitinase, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis.
To Be Published
|
|
7T1Q
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid | | Descriptor: | ACETATE ION, SUCCINIC ACID, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-02 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic Acid.
To be Published
|
|
4GIB
 
 | | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile | | Descriptor: | Beta-phosphoglucomutase, GLYCINE, PHOSPHATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile.
TO BE PUBLISHED
|
|
1SFS
 
 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
4OHJ
 
 | | Crystal structure of toxic shock syndrome toxin-1 (TSST-1) from Staphylococcus aureus | | Descriptor: | Toxic shock syndrome toxin-1 | | Authors: | Filippova, E.V, Shuvalova, L, Flores, K, Dubrovska, I, Bishop, B, Ngo, H, Yeats, C, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of toxic shock syndrome toxin-1 (TSST-1) from Staphylococcus aureus
To be Published
|
|
3LB0
 
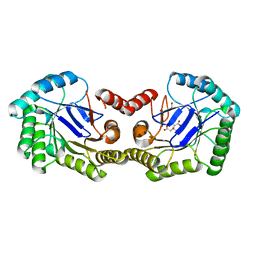 | | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-dehydroquinate dehydratase, CITRIC ACID, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Citrate Bound to the Active Site.
TO BE PUBLISHED
|
|
4ONX
 
 | | 2.8 Angstrom Crystal Structure of Sensor Domain of Histidine Kinase from Clostridium perfringens. | | Descriptor: | CHLORIDE ION, SULFATE ION, Sensor histidine kinase, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kwon, K, Shatsman, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-29 | | Release date: | 2014-03-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 Angstrom Crystal Structure of Sensor Domain of Histidine Kinase from Clostridium perfringens.
TO BE PUBLISHED
|
|
5VFB
 
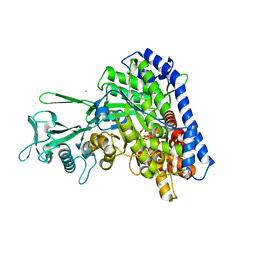 | | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid. | | Descriptor: | CHLORIDE ION, GLYCOLIC ACID, Malate synthase G, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.
To Be Published
|
|
3LAY
 
 | | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Zinc resistance-associated protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
5VGM
 
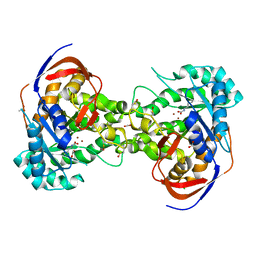 | | Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Lipowska, J, Shabalin, I.G, Miks, C.D, Winsor, J, Cooper, D.R, Shuvalova, L, Kwon, K, Lewinski, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-11 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
5VH6
 
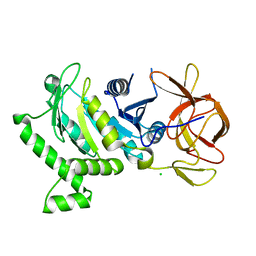 | | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis. | | Descriptor: | CHLORIDE ION, Elongation factor G | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-12 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-406) of Elongation Factor G from Bacillus subtilis.
To Be Published
|
|
8ER5
 
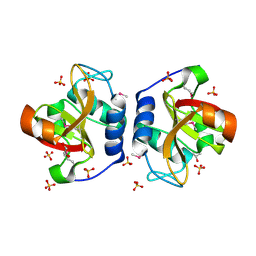 | | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NlpC/P60 domain-containing protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of NlpC/P60 domain from Clostridium innocuum NlpC/P60 domain-containing protein CI_01448.
To Be Published
|
|
1LVG
 
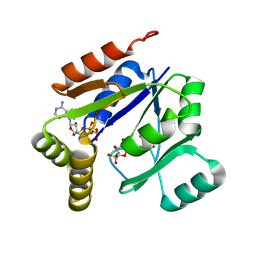 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | Authors: | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | Deposit date: | 2002-05-28 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
3BV6
 
 | | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold | | Descriptor: | FE (III) ION, Metal-dependent hydrolase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Yang, X, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold.
To be Published
|
|
6OV8
 
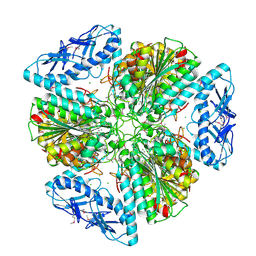 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6OAD
 
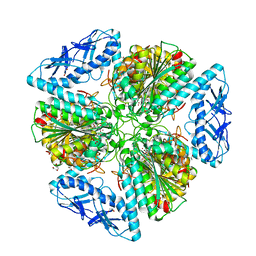 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
5DLL
 
 | | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Aminopeptidase N, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Borek, D, Raczynska, J, Dubrovska, I, Grimshaw, S, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Otwinowski, Z, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Aminopeptidase N (pepN) from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
3UWQ
 
 | | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Shuvalova, L, Kuhn, M, Filippova, E.V, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-02 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.80 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with uridine-5'-monophosphate (UMP)
To be Published
|
|
4G6X
 
 | | Crystal structure of glyoxalase/bleomycin resistance protein from Catenulispora acidiphila. | | Descriptor: | GLYCEROL, Glyoxalase/bleomycin resistance protein/dioxygenase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of glyoxalase/bleomycin resistance protein from Catenulispora acidiphila.
To be Published
|
|
3UPD
 
 | | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus | | Descriptor: | Ornithine carbamoyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Winsor, J, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | 2.9 Angstrom Crystal Structure of Ornithine Carbamoyltransferase (ArgF) from Vibrio vulnificus.
TO BE PUBLISHED
|
|
5DN8
 
 | | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP. | | Descriptor: | GTPase Der, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Minasov, G, Shuvalova, L, Han, A, Kim, H.-Y, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.76 Angstrom Crystal Structure of GTP-binding Protein Der from Coxiella burnetii in Complex with GDP.
To Be Published
|
|
4GFS
 
 | | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site | | Descriptor: | 3-dehydroquinate dehydratase, NICKEL (II) ION, SUCCINIC ACID | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 with Nickel Bound at Active Site
TO BE PUBLISHED
|
|
3LDV
 
 | | 1.77 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Glass, E.M, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | 1.77 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
9DUE
 
 | | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound SRM-07-081a | | Descriptor: | 3-chloro-6-(4-methylpiperazin-1-yl)-4-(pyridin-4-yl)pyridazine, BORIC ACID, Death-associated protein kinase 1, ... | | Authors: | Minasov, G, Winsor, J, Roy, S.M, Watterson, D.M, Shuvalova, L. | | Deposit date: | 2024-10-02 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound SRM-07-081a
To Be Published
|
|
