1EEJ
 
 | | CRYSTAL STRUCTURE OF THE PROTEIN DISULFIDE BOND ISOMERASE, DSBC, FROM ESCHERICHIA COLI | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, THIOL:DISULFIDE INTERCHANGE PROTEIN | | Authors: | McCarthy, A.A, Haebel, P.W, Torronen, A, Rybin, V, Baker, E.N, Metcalf, P. | | Deposit date: | 2000-01-31 | | Release date: | 2000-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli.
Nat.Struct.Biol., 7, 2000
|
|
2AYI
 
 | | Wild-type AmpT from Thermus thermophilus | | Descriptor: | Aminopeptidase T, ZINC ION | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Substrate Access to the Active Sites in Aminopeptidase T, a Representative of a New Metallopeptidase Clan.
J.Mol.Biol., 354, 2005
|
|
1ZJC
 
 | | Aminopeptidase S from S. aureus | | Descriptor: | COBALT (II) ION, aminopeptidase ampS | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Staphylococcus aureus Aminopeptidase S Is a Founding Member of a New Peptidase Clan.
J.Biol.Chem., 280, 2005
|
|
2JA9
 
 | | Structure of the N-terminal deletion of yeast exosome component Rrp40 | | Descriptor: | EXOSOME COMPLEX EXONUCLEASE RRP40 | | Authors: | Oddone, A, Lorentzen, E, Basquin, J, Gasch, A, Rybin, V, Conti, E, Sattler, M. | | Deposit date: | 2006-11-24 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Characterization of the Yeast Exosome Component Rrp40
Embo Rep., 8, 2007
|
|
5AHV
 
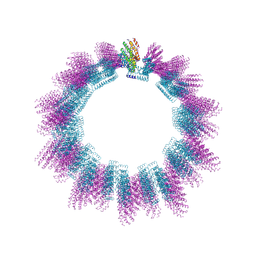 | | Cryo-EM structure of helical ANTH and ENTH tubules on PI(4,5)P2-containing membranes | | Descriptor: | ANTH DOMAIN OF ENDOCYTIC ADAPTOR SLA2, ENTH DOMAIN OF EPSIN ENT1 | | Authors: | Skruzny, M, Desfosses, A, Prinz, S, Dodonova, S.O, Gieras, A, Uetrecht, C, Jakobi, A.J, Abella, M, Hagen, W.J.H, Schulz, J, Meijers, R, Rybin, V, Briggs, J.A.G, Sachse, C, Kaksonen, M. | | Deposit date: | 2015-02-10 | | Release date: | 2015-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | An Organized Co-Assembly of Clathrin Adaptors is Essential for Endocytosis.
Dev.Cell, 33, 2015
|
|
4IQY
 
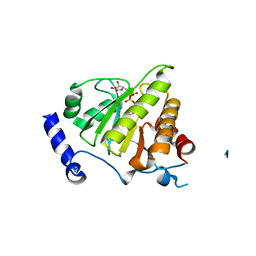 | | Crystal structure of the human protein-proximal ADP-ribosyl-hydrolase MacroD2 | | Descriptor: | MAGNESIUM ION, O-acetyl-ADP-ribose deacetylase MACROD2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Jankevicius, G, Hassler, M, Golia, B, Rybin, V, Zacharias, M, Timinszky, G, Ladurner, A.G. | | Deposit date: | 2013-01-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A family of macrodomain proteins reverses cellular mono-ADP-ribosylation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2LJX
 
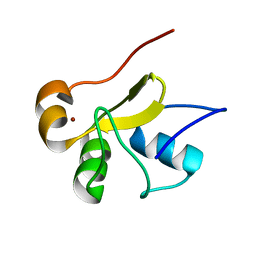 | | Structure of the monomeric N-terminal domain of HPV16 E6 oncoprotein | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
2LJZ
 
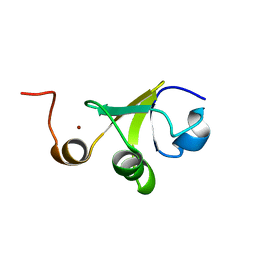 | | Structure of the C-terminal domain of HPV16 E6 oncoprotein | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
2NQC
 
 | | Crystal structure of ig-like domain 23 from human filamin C | | Descriptor: | Filamin-C, GLYCEROL, IMIDAZOLE, ... | | Authors: | Sjekloca, L, Pudas, R, Sjoeblom, B, Konarev, P, Carugo, O, Rybin, V, Kiema, T.R, Svergun, D, Ylanne, J, Djinovic-Carugo, K. | | Deposit date: | 2006-10-31 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23-24 dimer
J.Mol.Biol., 368, 2007
|
|
2LJY
 
 | | Haddock model structure of the N-terminal domain dimer of HPV16 E6 | | Descriptor: | Protein E6, ZINC ION | | Authors: | Zanier, K, Muhamed Sidi, A, Boulade-Ladame, C, Rybin, V, Chappelle, A, Atkinson, A, Kieffer, B, Trave, G. | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Analysis of the HPV16 E6 Oncoprotein Reveals a Self-Association Mechanism Required for E6-Mediated Degradation of p53.
Structure, 20, 2012
|
|
2Y8G
 
 | | Structure of the Ran-binding domain from human RanBP3 (E352A-R353V double mutant) | | Descriptor: | RAN-BINDING PROTEIN 3, SULFATE ION | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
2Y8F
 
 | | Structure of the Ran-binding domain from human RanBP3 (wild type) | | Descriptor: | CHLORIDE ION, GLYCEROL, RAN-BINDING PROTEIN 3 | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
2XQR
 
 | | Crystal structure of plant cell wall invertase in complex with a specific protein inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hothorn, M, Van den Ende, W, Lammens, W, Rybin, V, Scheffzek, K. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Insights Into the Ph-Controlled Targeting of Plant Cell-Wall Invertase by a Specific Inhibitor Protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2YN0
 
 | | tau55 histidine phosphatase domain | | Descriptor: | PHOSPHATE ION, TRANSCRIPTION FACTOR TAU 55 KDA SUBUNIT | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Iiic.
J.Biol.Chem., 288, 2013
|
|
2YN2
 
 | | Huf protein - paralogue of the tau55 histidine phosphatase domain | | Descriptor: | FORMIC ACID, UNCHARACTERIZED PROTEIN YNL108C | | Authors: | Taylor, N.M.I, Glatt, S, Hennrich, M, von Scheven, G, Grotsch, H, Fernandez-Tornero, C, Rybin, V, Gavin, A.C, Kolb, P, Muller, C.W. | | Deposit date: | 2012-10-11 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Functional Characterization of a Phosphatase Domain within Yeast General Transcription Factor Tfiiic.
J.Biol.Chem., 288, 2013
|
|
2VPB
 
 | | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | | Descriptor: | B-CELL CLL/LYMPHOMA 9 PROTEIN, PYGOPUS HOMOLOG 1, SODIUM ION, ... | | Authors: | Fiedler, M, Sanchez-Barrena, M.J, Nekrasov, M, Mieszczanek, J, Rybin, V, Muller, J, Evans, P, Bienz, M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Mol.Cell, 30, 2008
|
|
2VPG
 
 | | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | | Descriptor: | B-CELL CLL/LYMPHOMA 9 PROTEIN, GLYCEROL, HISTONE H3 TAIL, ... | | Authors: | Fiedler, M, Sanchez-Barrena, M.J, Nekrasov, M, Mieszczanek, J, Rybin, V, Muller, J, Bienz, M, Evans, P. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Mol.Cell, 30, 2008
|
|
2VPE
 
 | | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | | Descriptor: | B-CELL CLL/LYMPHOMA 9 PROTEIN, GLYCEROL, HISTONE H3 TAIL, ... | | Authors: | Fiedler, M, Sanchez-Barrena, M.J, Nekrasov, M, Mieszczanek, J, Rybin, V, Muller, J, Evans, P, Bienz, M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Mol.Cell, 30, 2008
|
|
2VP7
 
 | | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | | Descriptor: | B-CELL CLL/LYMPHOMA 9 PROTEIN, PYGOPUS HOMOLOG 1, SULFATE ION, ... | | Authors: | Fiedler, M, Sanchez-Barrena, M.J, Nekrasov, M, Mieszczanek, J, Rybin, V, Muller, J, Evans, P, Bienz, M. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Mol.Cell, 30, 2008
|
|
2VPD
 
 | | Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex | | Descriptor: | B-CELL CLL/LYMPHOMA 9 PROTEIN, PYGOPUS HOMOLOG 1, ZINC ION | | Authors: | Fiedler, M, Sanchez-Barrena, M.J, Nekrasov, M, Mieszczanek, J, Rybin, V, Muller, J, Evans, P, Bienz, M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Decoding of Methylated Histone H3 Tail by the Pygo- Bcl9 Wnt Signaling Complex.
Mol.Cell, 30, 2008
|
|
3CB6
 
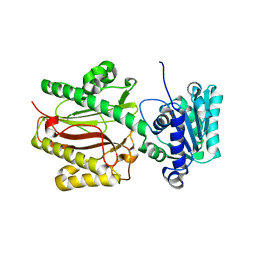 | | Crystal Structure of the S. pombe Peptidase Homology Domain of FACT complex subunit Spt16 (form B) | | Descriptor: | FACT complex subunit spt16 | | Authors: | Stuwe, T, Hothorn, M, Lejeune, E, Bortfeld-Miller, M, Scheffzek, K, Ladurner, A.G. | | Deposit date: | 2008-02-21 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The FACT Spt16 "peptidase" domain is a histone H3-H4 binding module
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2QUZ
 
 | |
3NSU
 
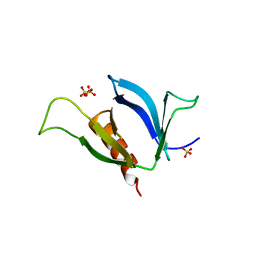 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
2BZT
 
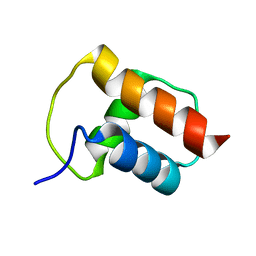 | | NMR structure of the bacterial protein YFHJ from E. coli | | Descriptor: | PROTEIN ISCX | | Authors: | Pastore, C, Kelly, G, Adinolfi, S, Mc Cormick, J.E, Pastore, A. | | Deposit date: | 2005-08-22 | | Release date: | 2006-12-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | YfhJ, a molecular adaptor in iron-sulfur cluster formation or a frataxin-like protein?
Structure, 14, 2006
|
|
5NEW
 
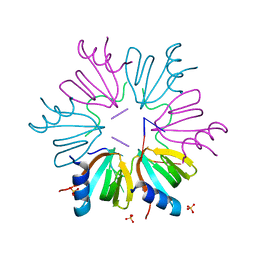 | |
