3SD3
 
 | | The structure of the tetrahydrofolate riboswitch containing a U25C mutation | | Descriptor: | IRIDIUM HEXAMMINE ION, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Tetrahydrofolate riboswitch | | Authors: | Reyes, F.E, Trausch, J.J, Ceres, P, Batey, R.T. | | Deposit date: | 2011-06-08 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer.
Structure, 19, 2011
|
|
4FRN
 
 | | Crystal structure of the cobalamin riboswitch regulatory element | | Descriptor: | BARIUM ION, Cobalamin riboswitch aptamer domain, Hydroxocobalamin | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4FRG
 
 | | Crystal structure of the cobalamin riboswitch aptamer domain | | Descriptor: | Hydroxocobalamin, IRIDIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
4GMA
 
 | | Crystal structure of the adenosylcobalamin riboswitch | | Descriptor: | Adenosylcobalamin, Adenosylcobalamin riboswitch | | Authors: | Reyes, F.E, Johnson, J.E, Polaski, J.T, Batey, R.T. | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | B12 cofactors directly stabilize an mRNA regulatory switch.
Nature, 492, 2012
|
|
3NPQ
 
 | |
3NPN
 
 | |
3GQU
 
 | |
3GQX
 
 | |
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | Descriptor: | IODIDE ION, Protein Ten1 | | Authors: | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5I9S
 
 | | MicroED structure of proteinase K at 1.75 A resolution | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Hattne, J, Shi, D, de la Cruz, M.J, Reyes, F.E, Gonen, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-08-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.75 Å) | | Cite: | Modeling truncated pixel values of faint reflections in MicroED images.
J.Appl.Crystallogr., 49, 2016
|
|
5O0U
 
 | | Crystal structure of tarantula venom peptide Protoxin-II | | Descriptor: | 1,2-ETHANEDIOL, Beta/omega-theraphotoxin-Tp2a, CHLORIDE ION | | Authors: | Tabor, A, McCarthy, S, Reyes, F.E. | | Deposit date: | 2017-05-17 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | The Role of Disulfide Bond Replacements in Analogues of the Tarantula Toxin ProTx-II and Their Effects on Inhibition of the Voltage-Gated Sodium Ion Channel Nav1.7.
J.Am.Chem.Soc., 139, 2017
|
|
5TY4
 
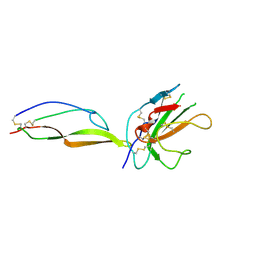 | | MicroED structure of a complex between monomeric TGF-b and its receptor, TbRII, at 2.9 A resolution | | Descriptor: | TGF-beta receptor type-2, mmTGF-b2-7m | | Authors: | Weiss, S.C, de la Cruz, M.J, Hattne, J, Shi, D, Reyes, F.E, Callero, G, Gonen, T. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.9 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
4OQU
 
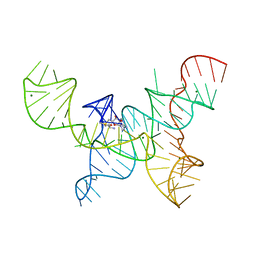 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92)) | | Descriptor: | MAGNESIUM ION, S-ADENOSYLMETHIONINE, SAM-I/IV riboswitch | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CSE
 
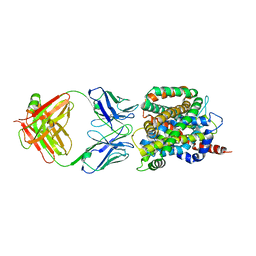 | | Crystal structure of sodium/alanine symporter AgcS with L-alanine bound | | Descriptor: | ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6DN2
 
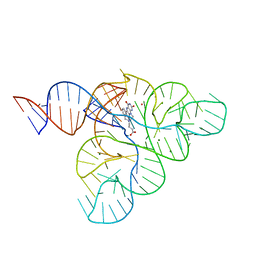 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1354 SPLIT RNA | | Descriptor: | 4-{benzyl[2-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)ethyl]amino}butanoic acid, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN3
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | Descriptor: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6DN1
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1151 SPLIT RNA | | Descriptor: | 10-(6-carboxyhexyl)-8-(cyclopentylamino)-2,4-dihydroxy-7-methylbenzo[g]pteridin-10-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6CSF
 
 | | Crystal structure of sodium/alanine symporter AgcS with D-alanine bound | | Descriptor: | D-ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4YEW
 
 | | HUab-19bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YF0
 
 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEX
 
 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFT
 
 | | HUab-20bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.914 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEY
 
 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
2XO1
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to N6-methyladenine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), Guanine riboswitch, ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
