7QRM
 
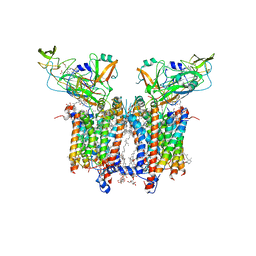 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.7 A resolution | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Sarewicz, M, Szwalec, M, Indyka, P, Rawski, M, Pintscher, S, Pietras, R, Mielecki, B, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
7PIK
 
 | | Cryo-EM structure of E. coli TnsB in complex with right end fragment of Tn7 transposon | | Descriptor: | Right end fragment of Tn7 transposon, Transposon Tn7 transposition protein TnsB | | Authors: | Kaczmarska, Z, Czarnocki-Cieciura, M, Rawski, M, Nowotny, M. | | Deposit date: | 2021-08-20 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis of transposon end recognition explains central features of Tn7 transposition systems.
Mol.Cell, 82, 2022
|
|
8Q3F
 
 | |
9ES8
 
 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f with decylplastoquinone bound at plastoquionol reduction site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES9
 
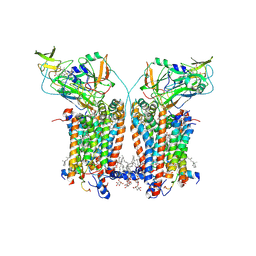 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with inhibitor DBMIB bound at plastoquinol oxidation site | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9ES7
 
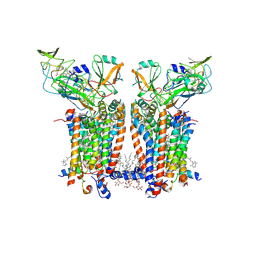 | | Cryo-EM structure of Spinacia oleracea cytochrome b6f complex with water molecules at 1.94 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Pietras, R, Pintscher, S, Mielecki, B, Szwalec, M, Wojcik-Augustyn, A, Indyka, P, Rawski, M, Koziej, L, Jaciuk, M, Wazny, G, Glatt, S, Osyczka, A. | | Deposit date: | 2024-03-25 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Molecular basis of plastoquinone reduction in plant cytochrome b 6 f.
Nat.Plants, 2024
|
|
9GRE
 
 | |
9GRD
 
 | |
7ZYV
 
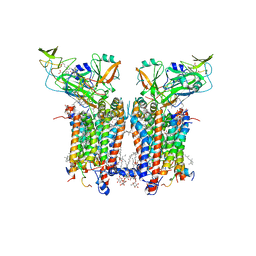 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
8AT6
 
 | | Cryo-EM structure of yeast Elp456 subcomplex | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASW
 
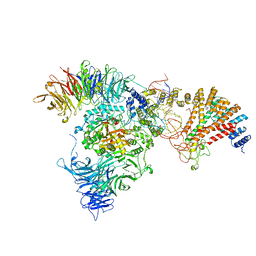 | | Cryo-EM structure of yeast Elp123 in complex with alanine tRNA | | Descriptor: | 5'-DEOXYADENOSINE, Alanine tRNA, Elongator complex protein 1, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8AVG
 
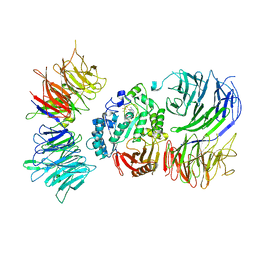 | | Cryo-EM structure of mouse Elp123 with bound SAM | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8ASV
 
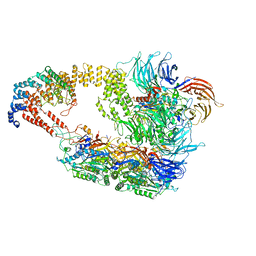 | | Cryo-EM structure of yeast Elongator complex | | Descriptor: | Elongator complex protein 1, Elongator complex protein 2, Elongator complex protein 3, ... | | Authors: | Jaciuk, M, Scherf, D, Kaszuba, K, Gaik, M, Koscielniak, A, Krutyholowa, R, Rawski, M, Indyka, P, Biela, A, Dobosz, D, Lin, T.-Y, Abbassi, N, Hammermeister, A, Chramiec-Glabik, A, Kosinski, J, Schaffrath, R, Glatt, S. | | Deposit date: | 2022-08-21 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the fully assembled Elongator complex.
Nucleic Acids Res., 51, 2023
|
|
8A0E
 
 | | CryoEM structure of DHS-eIF5A1 complex | | Descriptor: | Deoxyhypusine synthase, Eukaryotic translation initiation factor 5A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Wator, E, Wilk, P, Biela, A.P, Rawski, M, Grudnik, P. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
7OJ5
 
 | | Cryo-EM structure of Medicago truncatula HISN5 protein | | Descriptor: | Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Ruszkowski, M, Witek, W. | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAX
 
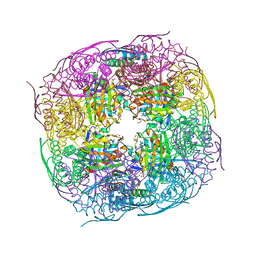 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, GOL and TRS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAV
 
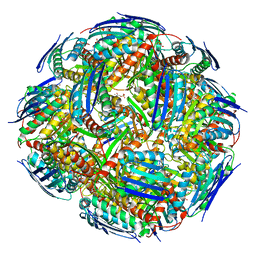 | | Medicago truncatula HISN5 (IGPD) in complex with MN and IG2 | | Descriptor: | (2S,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAY
 
 | | Medicago truncatula HISN5 (IGPD) in complex with MN, FMT, ACT, CIT, EDO, SO4 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CITRIC ACID, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
8QAW
 
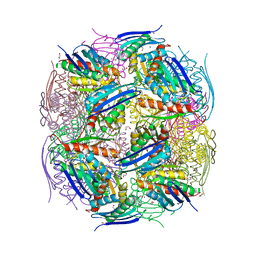 | | Medicago truncatula HISN5 (IGPD) in complex with MN, IMD, EDO, FMT, GOL and TRS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Witek, W, Ruszkowski, M. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Targeting imidazole-glycerol phosphate dehydratase in plants: novel approach for structural and functional studies, and inhibitor blueprinting.
Front Plant Sci, 15, 2024
|
|
7A6S
 
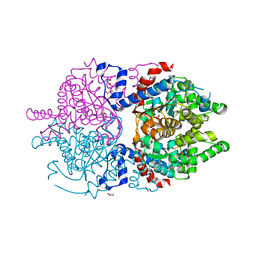 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-08-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
7A6T
 
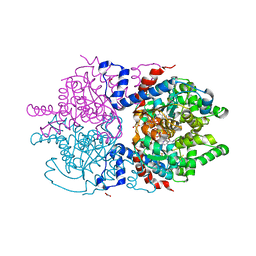 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase in complex with NAD and spermidine | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-08-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
8QO4
 
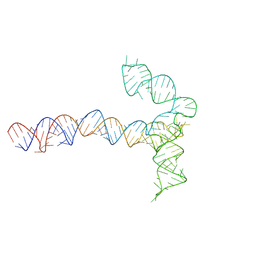 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | MERS-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO2
 
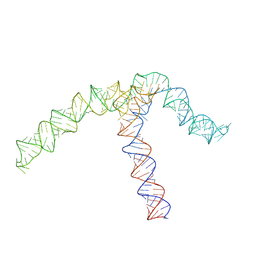 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | OC43-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO3
 
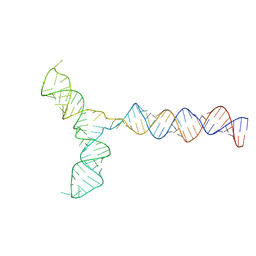 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | RoBat-CoV-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
8QO5
 
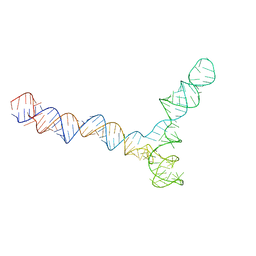 | | Conserved Structures and Dynamics in 5-Proximal Regions of Betacoronavirus RNA Genomes | | Descriptor: | SARS-CoV-2-SL5 | | Authors: | Moura, T.R, Purta, E, Bernat, A, Baulin, E, Mukherjee, S, Bujnicki, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.
Nucleic Acids Res., 52, 2024
|
|
