6MH3
 
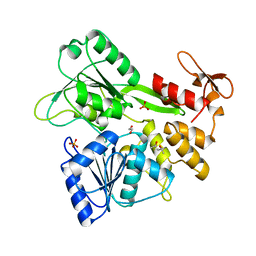 | | The crystal structure of Zika virus NS3 helicase domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Oliva, G, Mesquita, N, Godoy, A. | | Deposit date: | 2018-09-17 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of Zika virus NS3 helicase domain
To Be Published
|
|
1DEA
 
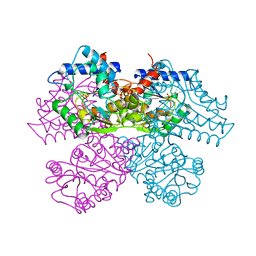 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1HOT
 
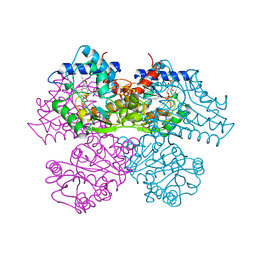 | | GLUCOSAMINE 6-PHOSPHATE DEAMINASE COMPLEXED WITH THE ALLOSTERIC ACTIVATOR N-ACETYL-GLUCOSAMINE-6-PHOSPHATE | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.L, Garratt, R, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-11-17 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1HOR
 
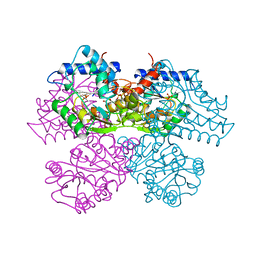 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1YRR
 
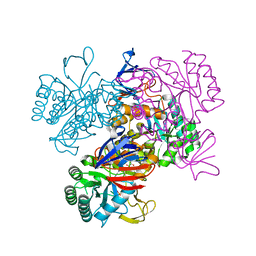 | | Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, PHOSPHATE ION | | Authors: | Ferreira, F.M, Aparicio, R, Mendoza-Hernandez, G, Calcagno, M.L, Oliva, G. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli.
J.Mol.Biol., 359, 2006
|
|
7N5Z
 
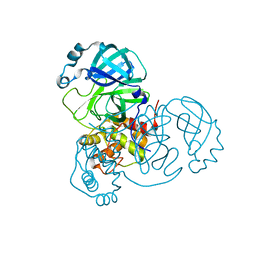 | | SARS-CoV-2 Main protease C145S mutant | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-06-07 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7N6N
 
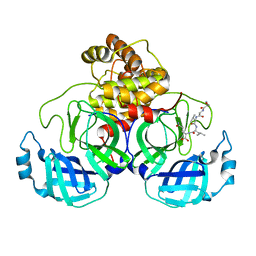 | | SARS-CoV-2 Main protease C145S mutant in complex with N and C-terminal residues | | Descriptor: | 3C-like proteinase | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7S82
 
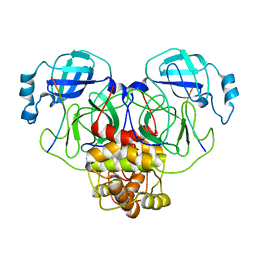 | |
1MVQ
 
 | | Cratylia mollis lectin (isoform 1) in complex with methyl-alpha-D-mannose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin, ... | | Authors: | de Souza, G.A, Oliveira, P.S, Trapani, S, Correia, M.T, Oliva, G, Coelho, L.C, Greene, L.J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Amino acid sequence and tertiary structure of Cratylia mollis seed lectin
Glycobiology, 13, 2003
|
|
1MZV
 
 | | Crystal Structure of Adenine Phosphoribosyltransferase (APRT) From Leishmania tarentolae | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine Phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Thiemann, O.H, Silva, M, Oliva, G, Silva, C.H.T.P, Iulek, J. | | Deposit date: | 2002-10-10 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Leishmania tarentolae: potential implications for APRT catalytic mechanism.
Biochim.Biophys.Acta, 1696, 2004
|
|
1SAC
 
 | | THE STRUCTURE OF PENTAMERIC HUMAN SERUM AMYLOID P COMPONENT | | Descriptor: | ACETIC ACID, CALCIUM ION, SERUM AMYLOID P COMPONENT | | Authors: | White, H.E, Emsley, J, O'Hara, B.P, Oliva, G, Srinivasan, N, Tickle, I.J, Blundell, T.L, Pepys, M.B, Wood, S.P. | | Deposit date: | 1994-01-27 | | Release date: | 1994-05-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of pentameric human serum amyloid P component.
Nature, 367, 1994
|
|
8UM3
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
6URV
 
 | | Crystal structure of Yellow Fever Virus NS2B-NS3 protease domain | | Descriptor: | NS2B, NS3 protease | | Authors: | Noske, G.D, Gawriljuk, V.F.O, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization and polymorphism analysis of the NS2B-NS3 protease from the 2017 Brazilian circulating strain of Yellow Fever virus.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
3OIS
 
 | | Crystal Structure Xylellain, a cysteine protease from Xylella fastidiosa | | Descriptor: | Cysteine protease, URIDINE-5'-DIPHOSPHATE | | Authors: | Leite, N.R, Faro, A.R, Oliva, M.A.V, Thiemann, O.H, Oliva, G. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of the cysteine protease Xylellain from Xylella fastidiosa reveals an intriguing activation mechanism.
Febs Lett., 587, 2013
|
|
8V7R
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
8V7U
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
1PZM
 
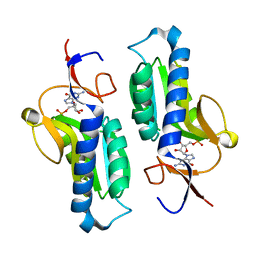 | | Crystal structure of HGPRT-ase from Leishmania tarentolae in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Monzani, P.S, Trapani, S, Oliva, G, Thiemann, O.H. | | Deposit date: | 2003-07-11 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Leishmania tarentolae hypoxanthine-guanine phosphoribosyltransferase.
Bmc Struct.Biol., 7, 2007
|
|
3IDS
 
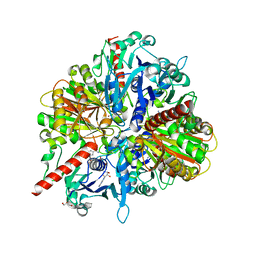 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetamide inhibitor | | Descriptor: | ACETAMIDE, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Balliano, T.L, Guido, R.V.C, Andricopulo, A.D, Oliva, G. | | Deposit date: | 2009-07-21 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetamide inhibitor
To be Published
|
|
3IEX
 
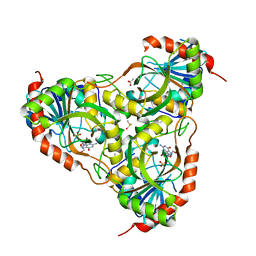 | | Schistosoma Purine nucleoside phosphorylase in complex with guanosine | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GUANOSINE, ... | | Authors: | Castilho, M.S, Pereira, H.M, Oliva, G, Andricopulo, A.D. | | Deposit date: | 2009-07-23 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for selective inhibition of purine nucleoside phosphorylase from Schistosoma mansoni: kinetic and structural studies
Bioorg.Med.Chem., 18, 2010
|
|
3K1E
 
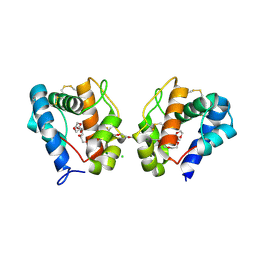 | | Crystal structure of odorant binding protein 1 (AaegOBP1) from Aedes aegypti | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Leite, N.R, Krogh, R, Leal, W.S, Iulek, J, Oliva, G. | | Deposit date: | 2009-09-27 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive "Lid".
Plos One, 4, 2009
|
|
2QED
 
 | | Crystal structure of Salmonella thyphimurium LT2 glyoxalase II | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Hydroxyacylglutathione hydrolase | | Authors: | Leite, N.R, Campos Bermudez, V.A, Krogh, R, Oliva, G, Soncini, F.C, Vila, A.J. | | Deposit date: | 2007-06-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of Salmonella typhimurium Glyoxalase II: New Insights into Metal Ion Selectivity
Biochemistry, 46, 2007
|
|
3PMP
 
 | | Crystal Structure of Cyclophilin A from Moniliophthora perniciosa in complex with Cyclosporin A | | Descriptor: | CYCLOSPORIN A, Cyclophilin A | | Authors: | Monzani, P, Pereira, H.M, Gramacho, K.P, Meirelles, F.V, Oliva, G, Cascardo, J.C.C. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Cyclophilin A from Moniliophthora perniciosa
To be Published
|
|
3O7T
 
 | | Crystal Structure of Cyclophilin A from Moniliophthora perniciosa | | Descriptor: | Cyclophilin A | | Authors: | Monzani, P.S, Pereira, H.M, Gramacho, K.P, Meirelles, F.V, Oliva, G, Cascardo, J.C.M. | | Deposit date: | 2010-07-31 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of apo-cyclophilin and bounded cyclosporine A from Moniliophthora perniciosa
To be Published
|
|
4E2B
 
 | | High resolution crystal structure of the old yellow enzyme from Trypanosoma cruzi | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Murakami, M.T, Rodrigues, N.C, Gava, L.M, Canduri, F, Oliva, G, Barbosa, L.R.S, Borgers, J.C. | | Deposit date: | 2012-03-08 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.269 Å) | | Cite: | High resolution crystal structure and in solution studies of the old yellow enzyme from Trypanosoma cruzi: Insights into oligomerization, enzyme dynamics and specificity
To be Published
|
|
1RP0
 
 | | Crystal Structure of Thi1 protein from Arabidopsis thaliana | | Descriptor: | ADENOSINE DIPHOSPHATE 5-(BETA-ETHYL)-4-METHYL-THIAZOLE-2-CARBOXYLIC ACID, HEPTANE-1,2,3-TRIOL, Thiazole biosynthetic enzyme, ... | | Authors: | Godoi, P.H.C, Van Sluys, M.A, Menck, C.F.M, Oliva, G. | | Deposit date: | 2003-12-02 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the thiazole biosynthetic enzyme THI1 from Arabidopsis thaliana.
J.Biol.Chem., 281, 2006
|
|
