1SRR
 
 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
1NAT
 
 | | CRYSTAL STRUCTURE OF SPOOF FROM BACILLUS SUBTILIS | | Descriptor: | SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Zapf, J, Hoch, J.A, Whiteley, J.M, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis.
Biochemistry, 36, 1997
|
|
1L3R
 
 | | Crystal Structure of a Transition State Mimic of the Catalytic Subunit of cAMP-dependent Protein Kinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Madhusudan, Akamine, P, Xuong, N.-H, Taylor, S.S. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase.
Nat.Struct.Biol., 9, 2002
|
|
1LMA
 
 | |
1JBP
 
 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Substrate Peptide, ADP and Detergent | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, MUSCLE/BRAIN FORM, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
1JLU
 
 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
1J3H
 
 | | Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Wu, J, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic Features of cAMP-dependent Protein Kinase Revealed by Apoenzyme Crystal Structure
J.Mol.Biol., 327, 2003
|
|
1SYK
 
 | | Crystal structure of E230Q mutant of cAMP-dependent protein kinase reveals unexpected apoenzyme conformation | | Descriptor: | cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Wu, J, Yang, J, Madhusudan, N, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2004-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the E230Q mutant of cAMP-dependent
protein kinase reveals an unexpected apoenzyme conformation and an
extended N-terminal A helix.
Protein Sci., 14, 2005
|
|
1F51
 
 | | A TRANSIENT INTERACTION BETWEEN TWO PHOSPHORELAY PROTEINS TRAPPED IN A CRYSTAL LATTICE REVEALS THE MECHANISM OF MOLECULAR RECOGNITION AND PHOSPHOTRANSFER IN SINGAL TRANSDUCTION | | Descriptor: | MAGNESIUM ION, SPORULATION INITIATION PHOSPHOTRANSFERASE B, SPORULATION INITIATION PHOSPHOTRANSFERASE F | | Authors: | Zapf, J, Sen, U, Madhusudan, M, Hoch, J.A, Varughese, K.I. | | Deposit date: | 2000-06-11 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A transient interaction between two phosphorelay proteins trapped in a crystal lattice reveals the mechanism of molecular recognition and phosphotransfer in signal transduction.
Structure Fold.Des., 8, 2000
|
|
1REJ
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-HYDROXYBENZOATE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1REK
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 8 | | Descriptor: | 3-[(3-SEC-BUTYL-4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXY-5-METHOXYBENZOYL)BENZOATE, PENTANAL, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1RE8
 
 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 2 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXYBENZOYL)BENZOATE, N-OCTANE, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1KGZ
 
 | | Crystal Structure Analysis of the Anthranilate Phosphoribosyltransferase from Erwinia carotovora (current name, Pectobacterium carotovorum) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Anthranilate phosphoribosyltransferase, MANGANESE (II) ION, ... | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-28 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
1KHD
 
 | | Crystal Structure Analysis of the anthranilate phosphoribosyltransferase from Erwinia carotovora at 1.9 resolution (current name, Pectobacterium carotovorum) | | Descriptor: | Anthranilate phosphoribosyltransferase | | Authors: | Kim, C, Xuong, N.-H, Edwards, S, Madhusudan, Yee, M.-C, Spraggon, G, Mills, S.E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-10-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Crystal Structure of Anthranilate
Phosphoribosyltransferase from the
Enterobacterium Pectobacterium carotovorum
FEBS Lett., 523, 2002
|
|
1LQ1
 
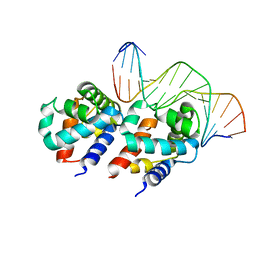 | | DNA Complexed Structure of the Key Transcription Factor Initiating Development in Sporulation Bacteria | | Descriptor: | 5'-D(*AP*CP*AP*AP*AP*AP*TP*TP*CP*GP*AP*CP*AP*CP*GP*A)-3', 5'-D(*TP*TP*CP*GP*TP*GP*TP*CP*GP*AP*AP*TP*TP*TP*TP*G)-3', Stage 0 sporulation protein A | | Authors: | Zhao, H, Msadek, T, Zapf, J, Madhusudan, Hoch, J.A, Varughese, K.I. | | Deposit date: | 2002-05-08 | | Release date: | 2002-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA complexed structure of the key transcription factor initiating development in sporulating bacteria.
Structure, 10, 2002
|
|
1OAK
 
 | |
1CX4
 
 | |
1RDQ
 
 | | Hydrolysis of ATP in the crystal of Y204A mutant of cAMP-dependent protein kinase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Ten Eyck, L.F, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-05 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a cAMP-dependent Protein Kinase Mutant at 1.26A: New Insights into the Catalytic Mechanism.
J.Mol.Biol., 336, 2004
|
|
1RHA
 
 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1RHB
 
 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1IXM
 
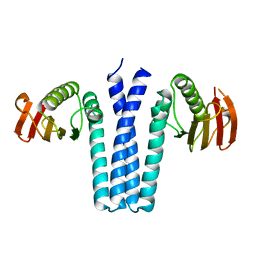 | | CRYSTAL STRUCTURE OF SPOOB FROM BACILLUS SUBTILIS | | Descriptor: | PROTEIN (SPORULATION RESPONSE REGULATORY PROTEIN) | | Authors: | Varughese, K.I. | | Deposit date: | 1998-11-06 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase.
Mol.Cell, 2, 1998
|
|
1UCO
 
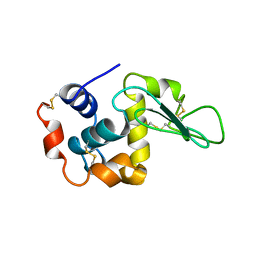 | | HEN EGG-WHITE LYSOZYME, LOW HUMIDITY FORM | | Descriptor: | LYSOZYME | | Authors: | Nagendra, H.G, Sudarsanakumar, C, Vijayan, M. | | Deposit date: | 1995-12-31 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An X-ray analysis of native monoclinic lysozyme. A case study on the reliability of refined protein structures and a comparison with the low-humidity form in relation to mobility and enzyme action.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1XEK
 
 | |
1XEI
 
 | |
1XEJ
 
 | |
