197D
 
 | |
1D58
 
 | | THE MOLECULAR STRUCTURE OF A 4'-EPIADRIAMYCIN COMPLEX WITH D(TGATCA) AT 1.7 ANGSTROM RESOLUTION-COMPARISON WITH THE STRUCTURE OF 4'-EPIADRIAMYCIN D(TGTACA) AND D(CGATCG) COMPLEXES | | Descriptor: | 4'-EPIDOXORUBICIN, DNA (5'-D(*TP*GP*AP*TP*CP*A)-3') | | Authors: | Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-02-20 | | Release date: | 1992-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular structure of a 4'-epiadriamycin complex with d(TGATCA) at 1.7A resolution: comparison with the structure of 4'-epiadriamycin d(TGTACA) and d(CGATCG) complexes.
Nucleic Acids Res., 20, 1992
|
|
1R03
 
 | | crystal structure of a human mitochondrial ferritin | | Descriptor: | MAGNESIUM ION, mitochondrial ferritin | | Authors: | Corsi, B, Santambrogio, P, Arosio, P, Levi, S, Langlois d'Estaintot, B, Granier, T, Gallois, B, Chevallier, J.M, Precigoux, G. | | Deposit date: | 2003-09-19 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure and Biochemical Properties of the Human Mitochondrial Ferritin and its Mutant Ser144Ala
J.Mol.Biol., 340, 2004
|
|
4NH4
 
 | | Structure of the binary complex of a zingiber officinale double bond reductase in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Zingiber officinale double bond reductase | | Authors: | Langlois D'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2013-11-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of zingiber officinale double bond reductase
To be Published
|
|
2H0L
 
 | | Crystal Structure of a Mutant of Rat Annexin A5 | | Descriptor: | Annexin A5, CALCIUM ION | | Authors: | Langlois D'Estaintot, B, Gallois, B, Granier, T, Tessier, B, Brisson, A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Identification of the Residues Involved in the Formation of Annexin V Trimers within 2D and 3D Crystals
To be Published
|
|
2IE6
 
 | | Annexin V under 2.0 MPa pressure of xenon | | Descriptor: | Annexin A5, CALCIUM ION, SULFATE ION, ... | | Authors: | Colloc'h, N, Sopkova-de Oliveira Santos, J, Prange, T, Brisson, A, Langlois d'Estainto, B, Gallois, B. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Protein Crystallography under Xenon and Nitrous Oxide Pressure: Comparison with In Vivo Pharmacology Studies and Implications for the Mechanism of Inhaled Anesthetic Action
Biophys.J., 92, 2007
|
|
2IE7
 
 | | Annexin V under 2.0 MPa pressure of nitrous oxide | | Descriptor: | Annexin A5, CALCIUM ION, NITROUS OXIDE, ... | | Authors: | Colloc'h, N, Sopkova-de Oliveira Santos, J, Prange, T, Brisson, A, Langlois d'Estainto, B, Gallois, B. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein Crystallography under Xenon and Nitrous Oxide Pressure: Comparison with In Vivo Pharmacology Studies and Implications for the Mechanism of Inhaled Anesthetic Action
Biophys.J., 92, 2007
|
|
4LP6
 
 | | Crystal Structure of Human Carbonic Anhydrase II in complex with a quinoline oligoamide foldamer | | Descriptor: | 8-({[4-(3-aminopropoxy)-8-({[4-hydroxy-8-({[4-(2-methylpropoxy)-8-({[4-(3-{[(4-sulfamoylbenzoyl)amino]methyl}phenoxy)butyl]carbamoyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)-4-(carboxymethoxy)quinoline-2-carboxylic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Buratto, J, Granier, T, Langlois D'estaintot, B, Huc, I, Gallois, B. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 angstrom resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1KRQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CAMPYLOBACTER JEJUNI FERRITIN | | Descriptor: | ferritin | | Authors: | Hortolan, L, Saintout, N, Granier, G, Langlois d'Estaintot, B, Manigand, C, Mizunoe, Y, Wai, S.N, Gallois, B, Precigoux, G. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURE OF CAMPYLOBACTER JEJUNI FERRITIN AT 2.7 A RESOLUTION
To be Published
|
|
2IOD
 
 | | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Dihydroflavonol 4-reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Langlois d'Estaintot, B, Granier, T, Gallois, B. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site
To be Published
|
|
4WE3
 
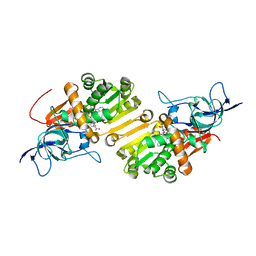 | | STRUCTURE OF THE BINARY COMPLEX OF A ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE IN COMPLEX WITH NADP MONOCLINIC CRYSTAL FORM | | Descriptor: | Double Bond Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Collery, J, Langlois d'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2014-09-09 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | STRUCTURE OF ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE
to be published
|
|
4WGG
 
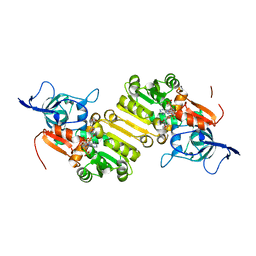 | | STRUCTURE OF THE TERNARY COMPLEX OF A ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE IN COMPLEX WITH NADP AND CONIFERYL ALDEHYDE | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, Double Bond Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Collery, J, Langlois d'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2014-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE
to be published
|
|
1IES
 
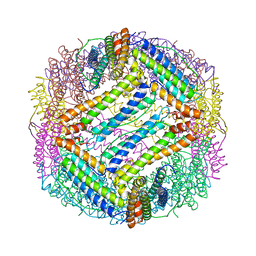 | | TETRAGONAL CRYSTAL STRUCTURE OF NATIVE HORSE SPLEEN FERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Granier, T, Gallois, B, Dautant, A, Langlois D'Estaintot, B, Precigoux, G. | | Deposit date: | 1996-05-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of the structures of the cubic and tetragonal forms of horse-spleen apoferritin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
5HIX
 
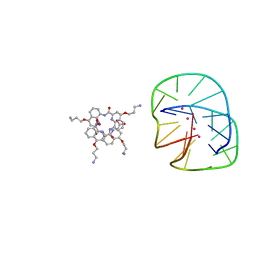 | | Cocrystal structure of an anti-parallel DNA G-quadruplex and a tetra-Quinoline Foldamer | | Descriptor: | 4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-nitroquinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinoline-2-carboxylic acid, Dimeric G-quadruplex, POTASSIUM ION | | Authors: | Mandal, P.K, Baptiste, B, Langlois d'Estaintot, B, Kauffmann, B, Huc, I. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multivalent Interactions between an Aromatic Helical Foldamer and a DNA G-Quadruplex in the Solid State.
Chembiochem, 17, 2016
|
|
1IER
 
 | | CUBIC CRYSTAL STRUCTURE OF NATIVE HORSE SPLEEN FERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Granier, T, Gallois, B, Dautant, A, Langlois D'Estaintot, B, Precigoux, G. | | Deposit date: | 1996-05-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of the structures of the cubic and tetragonal forms of horse-spleen apoferritin.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
6HZX
 
 | | Protein-aromatic foldamer complex crystal structure | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Post, S, Langlois d'Estaintot, B, Fischer, L, Granier, T, Huc, I. | | Deposit date: | 2018-10-24 | | Release date: | 2019-09-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure Elucidation of Helical Aromatic Foldamer-Protein Complexes with Large Contact Surface Areas.
Chemistry, 25, 2019
|
|
1LB3
 
 | | Structure of recombinant mouse L chain ferritin at 1.2 A resolution | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, GLYCEROL, ... | | Authors: | Granier, T, Langlois D'Estaintot, B, Gallois, B, Chevalier, J.-M, Precigoux, G, Santambrogio, P, Arosio, P. | | Deposit date: | 2002-04-02 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural description of the active sites of mouse L-chain ferritin at 1.2A resolution
J.Biol.Inorg.Chem., 8, 2003
|
|
2RH8
 
 | | Structure of apo anthocyanidin reductase from vitis vinifera | | Descriptor: | Anthocyanidin reductase, CHLORIDE ION | | Authors: | Gargouri, M, Mauge, C, Langlois D'Estaintot, B, Granier, T, Manigan, C, Gallois, B. | | Deposit date: | 2007-10-08 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3HFS
 
 | | Structure of apo anthocyanidin reductase from vitis vinifera | | Descriptor: | Anthocyanidin reductase, CHLORIDE ION | | Authors: | Gargouri, M, Mauge, C, Langlois d'Estaintot, B, Granier, T, Manigan, C, Gallois, B. | | Deposit date: | 2009-05-12 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5LVS
 
 | | Self-assembled protein-aromatic foldamer complexes with 2:3 and 2:2:1 stoichiometries | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Jewginski, M, LANGLOIS D'ESTAINTOT, B, Granier, T, Huc, Y. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
28DN
 
 | | CRYSTAL STRUCTURE ANALYSIS OF AN A(DNA) OCTAMER D(GTACGTAC) | | Descriptor: | DNA (5'-D(*GP*TP*AP*CP*GP*TP*AP*C)-3') | | Authors: | Courseille, C, Dautant, A, Hospital, M, Langlois D'Estaintot, B, Precigoux, G, Molko, D, Teoule, R. | | Deposit date: | 1990-05-03 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of an A(DNA) Octamer d(GTACGTAC)
Acta Crystallogr.,Sect.A, 46, 1990
|
|
198D
 
 | | A TRIGONAL FORM OF THE IDARUBICIN-D(CGATCG) COMPLEX: CRYSTAL AND MOLECULAR STRUCTURE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), IDARUBICIN, SPERMINE | | Authors: | Dautant, A, Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1994-11-28 | | Release date: | 1995-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A trigonal form of the idarubicin:d(CGATCG) complex; crystal and molecular structure at 2.0 A resolution.
Nucleic Acids Res., 23, 1995
|
|
3C1T
 
 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
3BXX
 
 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-15 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
3KXU
 
 | | Crystal structure of human ferritin FTL498InsTC pathogenic mutant | | Descriptor: | CADMIUM ION, Ferritin, SULFATE ION | | Authors: | Granier, T, Gallois, B, Langlois d'Estaintot, B, Arosio, P. | | Deposit date: | 2009-12-04 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mutant ferritin L-chains that cause neurodegeneration act in a dominant-negative manner to reduce ferritin iron incorporation.
J.Biol.Chem., 285, 2010
|
|
