7ABW
 
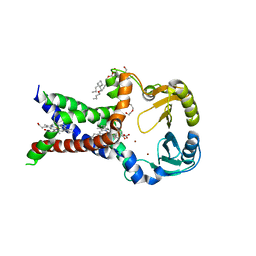 | | Crystal structure of siderophore reductase FoxB | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DECYL-BETA-D-MALTOPYRANOSIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2020-09-09 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural insights into a novel family of integral membrane siderophore reductases.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5EDL
 
 | | Crystal structure of an S-component of ECF transporter | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Putative HMP/thiamine permease protein YkoE, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2015-10-21 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a Group I Energy Coupling Factor Vitamin Transporter S Component in Complex with Its Cognate Substrate.
Cell Chem Biol, 23, 2016
|
|
6I96
 
 | |
6I98
 
 | |
6I97
 
 | |
6Z8A
 
 | | Outer membrane FoxA in complex with nocardamine | | Descriptor: | (8E)-6,17,28-trihydroxy-1,6,12,17,23,28-hexaazacyclotritriacont-8-ene-2,5,13,16,24,27-hexone, DIMETHYL SULFOXIDE, FE (III) ION, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2020-06-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Nocardamine-Dependent Iron Uptake in Pseudomonas aeruginosa : Exclusive Involvement of the FoxA Outer Membrane Transporter.
Acs Chem.Biol., 15, 2020
|
|
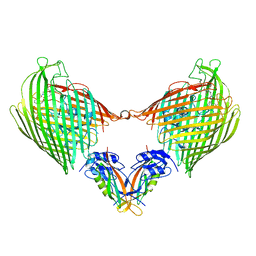
5VTG
 
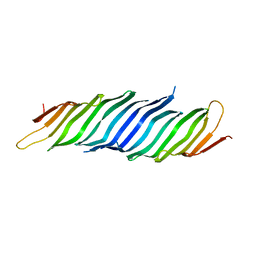 | |
8B43
 
 | | Crystal structure of ferrioxamine transporter | | Descriptor: | 1,12-bis(oxidanyl)-1,6,12,17-tetrazacyclodocosane-2,5,13,16-tetrone, FE (III) ION, Ferrichrome-iron receptor, ... | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2022-09-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Interactions of TonB-dependent transporter FoxA with siderophores and antibiotics that affect binding, uptake, and signal transduction.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5NOH
 
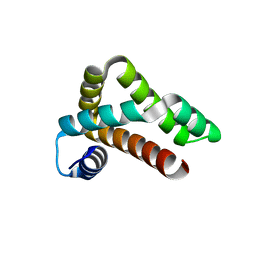 | | HRSV M2-1 core domain | | Descriptor: | Transcription elongation factor M2-1 | | Authors: | Josts, I, Almeida Hernandez, Y, Molina, I.G, de Prat-Gay, G, Tidow, H. | | Deposit date: | 2017-04-12 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4RTD
 
 | | Escherichia coli alpha-2-macroglobulin activated by porcine elastase | | Descriptor: | Uncharacterized lipoprotein YfhM | | Authors: | Fyfe, C.D, Grinter, R, Roszak, A.W, Josts, I, Cogdell, R.J, Walker, D. | | Deposit date: | 2014-11-14 | | Release date: | 2015-07-15 | | Last modified: | 2015-07-29 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structure of protease-cleaved Escherichia coli alpha-2-macroglobulin reveals a putative mechanism of conformational activation for protease entrapment.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5NKX
 
 | | HRSV M2-1 core domain, P3221 crystal form | | Descriptor: | M2-1 | | Authors: | Almeida Hernandez, Y, Josts, I, Molina, I.G, de Pray-Gay, G, Tidow, H. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.00008678 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4ZHO
 
 | | The crystal structure of Arabidopsis ferredoxin 2 with 2Fe-2S cluster | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZHP
 
 | | The crystal structure of Potato ferredoxin I with 2Fe-2S cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Potato Ferredoxin I | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4ZGV
 
 | | The Crystal Structure of the Ferredoxin Receptor FusA from Pectobacterium atrosepticum SCRI1043 | | Descriptor: | Ferredoxin receptor, LAURYL DIMETHYLAMINE-N-OXIDE, octyl beta-D-glucopyranoside | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2015-04-24 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacterial plant-ferredoxin receptor FusA.
Nat Commun, 7, 2016
|
|
4QKO
 
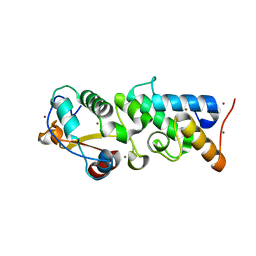 | | The Crystal Structure of the Pyocin S2 Nuclease Domain, Immunity Protein Complex at 1.8 Angstroms | | Descriptor: | BROMIDE ION, MAGNESIUM ION, Pyocin-S2, ... | | Authors: | Grinter, R, Josts, I, Roszak, A.W, Cogdell, C.J, Walker, D. | | Deposit date: | 2014-06-07 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into pyocin S2
To be Published
|
|
6HCS
 
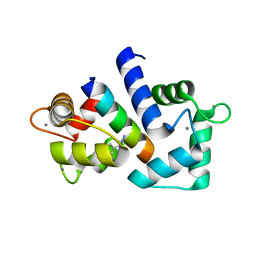 | |
4QAY
 
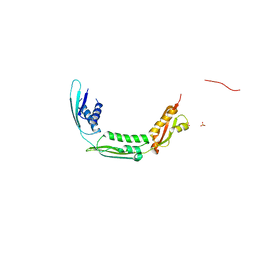 | |
7BCW
 
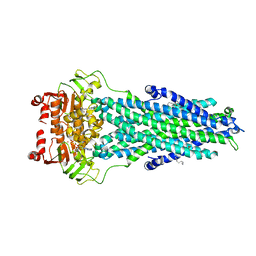 | | Structure of MsbA in Salipro with ADP vanadate | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent lipid A-core flippase, ... | | Authors: | Traore, D.A.K, Tidow, H. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of MsbA in saposin-lipid nanoparticles (Salipro) provides insights into nucleotide coordination.
Febs J., 289, 2022
|
|
7MHU
 
 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
6H79
 
 | |
4UHP
 
 | | Crystal structure of the pyocin AP41 DNase-Immunity complex | | Descriptor: | BACTERIOCIN IMMUNITY PROTEIN, LARGE COMPONENT OF PYOCIN AP41 | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
4UHQ
 
 | | Crystal structure of the pyocin AP41 DNase | | Descriptor: | CITRIC ACID, LARGE COMPONENT OF PYOCIN AP41, NICKEL (II) ION | | Authors: | Joshi, A, Chen, S, Wojdyla, J.A, Kaminska, R, Kleanthous, C. | | Deposit date: | 2015-03-25 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of the Ultra-High Affinity Protein-Protein Complexes of Pyocins S2 and Ap41 and Their Cognate Immunity Proteins from Pseudomonas Aeruginosa
J.Mol.Biol., 427, 2015
|
|
5EW5
 
 | |
4N58
 
 | | Crystal Structure of Pectocin M2 at 1.86 Angstroms | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Grinter, R, Roszak, A.W, Zeth, K, Cogdell, C.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
4N59
 
 | | The Crystal Structure of Pectocin M2 at 2.3 Angstroms | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Pectocin M2, ... | | Authors: | Zeth, K, Grinter, R, Roszak, A.W, Cogdell, R.J, Walker, D. | | Deposit date: | 2013-10-09 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake.
Mol.Microbiol., 93, 2014
|
|
