6SE5
 
 | |
6SE7
 
 | |
8CAT
 
 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
7CAT
 
 | | The NADPH binding site on beef liver catalase | | Descriptor: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | Deposit date: | 1984-11-15 | | Release date: | 1985-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
3D6E
 
 | | Crystal structure of the engineered 1,3-1,4-beta-glucanase protein from Bacillus licheniformis | | Descriptor: | Beta-glucanase, CALCIUM ION | | Authors: | Fita, I, Planas, A, Calisto, B.M, Addington, T. | | Deposit date: | 2008-05-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Re-engineering specificity in 1,3-1,4-beta-glucanase to accept branched xyloglucan substrates
Proteins, 79, 2011
|
|
1QGC
 
 | |
6RUT
 
 | |
6S3U
 
 | | Adhesin P140 from Mycoplasma Genitalium | | Descriptor: | Adhesin P1 | | Authors: | Fita, I, Aparicio, D. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structure and mechanism of the Nap adhesion complex from the human pathogen Mycoplasma genitalium.
Nat Commun, 11, 2020
|
|
6TM0
 
 | | N-Domain P40/P90 Mycoplasma pneumoniae complexed with 6'SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Vizarraga, D, Aparicio, D, Illanes, R, Fita, I, Perez-Luque, R, Martin, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae.
Nat Commun, 11, 2020
|
|
1CF9
 
 | |
1A3R
 
 | |
2V5H
 
 | | Controlling the storage of nitrogen as arginine: the complex of PII and acetylglutamate kinase from Synechococcus elongatus PCC 7942 | | Descriptor: | ACETYLGLUTAMATE KINASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Llacer, J.L, Marco-Marin, C, Gil-Ortiz, F, Fita, I, Rubio, V. | | Deposit date: | 2007-07-04 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Crystal Structure of the Complex of Pii and Acetylglutamate Kinase Reveals How Pii Controls the Storage of Nitrogen as Arginine.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
8ROR
 
 | | Single-particle cryo-EM of Mycoplasma pneumoniae adhesin P1 complexed with the anti-adhesive Fab fragment. | | Descriptor: | Adhesin P1, Heavy Chain Fab, Light Chain Fab | | Authors: | Vizarraga, D, Kawamoto, A, Marcos-Silva, M, Fita, I, Miyata, M, Pinyol, J, Namba, K, Kenri, T. | | Deposit date: | 2024-01-12 | | Release date: | 2025-03-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Dynamics of the adhesion complex of the human pathogens Mycoplasma pneumoniae and Mycoplasma genitalium.
Plos Pathog., 21, 2025
|
|
1GGE
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, NATIVE STRUCTURE AT 1.9 A RESOLUTION. | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, PROTEIN (CATALASE HPII) | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-16 | | Release date: | 2000-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1GGJ
 
 | | CRYSTAL STRUCTURE OF CATALASE HPII FROM ESCHERICHIA COLI, ASN201ALA VARIANT. | | Descriptor: | CATALASE HPII, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE | | Authors: | Melik-Adamyan, W.R, Bravo, J, Carpena, X, Switala, J, Mate, M.J, Fita, I, Loewen, P.C. | | Deposit date: | 2000-08-21 | | Release date: | 2000-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate flow in catalases deduced from the crystal structures of active site variants of HPII from Escherichia coli.
Proteins, 44, 2001
|
|
1IPH
 
 | |
4XNG
 
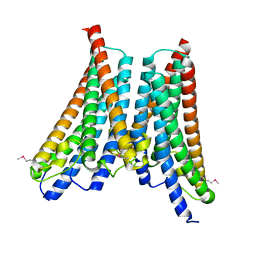 | |
6F2W
 
 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | ALPHA-AMINOISOBUTYRIC ACID, Nanobody 74, Putative amino acid/polyamine transport protein, ... | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
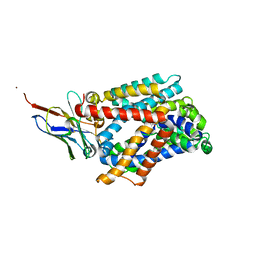
6F2G
 
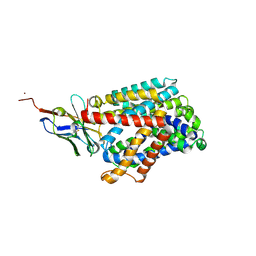 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | Nanobody 74, Putative amino acid/polyamine transport protein, ZINC ION | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
3L01
 
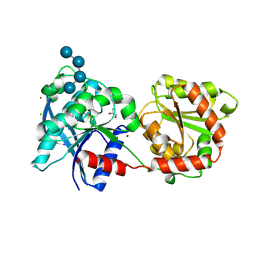 | | Crystal structure of monomeric glycogen synthase from Pyrococcus abyssi | | Descriptor: | CHLORIDE ION, GLYCEROL, GlgA glycogen synthase, ... | | Authors: | Diaz, A, Martinez-Pons, C, Fita, I, Ferrer, J.C, Guinovart, J.J. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Processivity and Subcellular Localization of Glycogen Synthase Depend on a Non-catalytic High Affinity Glycogen-binding Site.
J.Biol.Chem., 286, 2011
|
|
4RPE
 
 | |
1U2L
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P1) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2J
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P21 21 21) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2K
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (I41) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1BBD
 
 | |
