5WX9
 
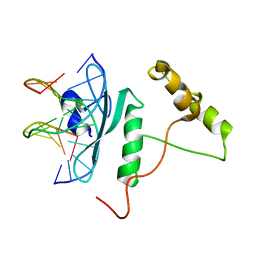 | | Crystal Structure of AtERF96 with GCC-box | | Descriptor: | Ethylene-responsive transcription factor ERF096, GCC-box motif | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2017-01-06 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into Arabidopsis ethylene response factor 96 with an extended N-terminal binding to GCC box.
Plant Mol.Biol., 104, 2020
|
|
5ECQ
 
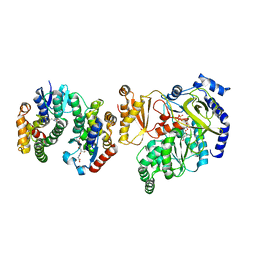 | | Crystal Structure of FIN219-FIP1 complex with JA, VAL and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECN
 
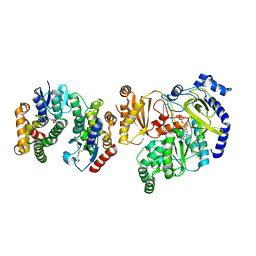 | | Crystal Structure of FIN219-FIP1 complex with JA, Leu and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECM
 
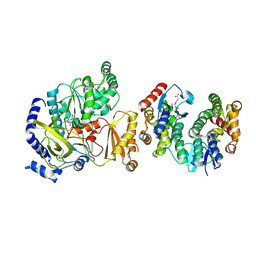 | | Crystal Structure of FIN219-FIP1 complex with JA and Leu | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECO
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, Leu and Mg | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECH
 
 | | Crystal Structure of FIN219-FIP1 complex with JA and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECK
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, Ile and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECL
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, Ile and Mg | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, ISOLEUCINE, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECI
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, ATP and Mg | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECR
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, VAL and Mg | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECP
 
 | | Crystal Structure of FIN219-FIP1 complex with JA, MET and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25001574 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECS
 
 | | Crystal Structure of FIP1 with GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20 | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6500026 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5GZZ
 
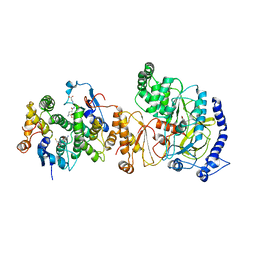 | | Crystal Structure of FIN219-SjGST complex with JA | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6WQE
 
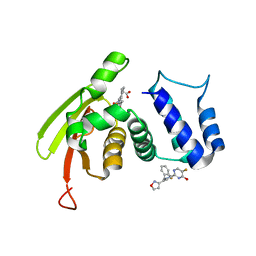 | | Solution Structure of the IWP-051-bound H-NOX from Shewanella woodyi in the Fe(II)CO ligation state | | Descriptor: | 5-fluoro-2-{1-[(2-fluorophenyl)methyl]-5-(1,2-oxazol-3-yl)-1H-pyrazol-3-yl}pyrimidin-4-ol, CARBON MONOXIDE, Heme NO binding domain protein, ... | | Authors: | Chen, C.Y, Lee, W, Montfort, W.R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051.
Protein Sci., 30, 2021
|
|
6KPE
 
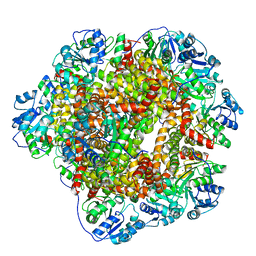 | | 343 K cryoEM structure of Sso-KARI in complex with Mg2+ | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KQJ
 
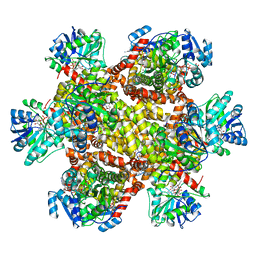 | | 309 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KOU
 
 | | 277 K cryoEM structure of Sso-KARI in complex with magnesium ions | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KQ4
 
 | | 323 K cryoEM structure of Sso-KARI in complex with Mg2+ | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-16 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KPI
 
 | | 298 K cryoEM structure of Sso-KARI in complex with Mg2+ | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KPJ
 
 | | 298 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KQ8
 
 | | 328 K cryoEM structure of Sso-KARI in complex with Mg2+ | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-16 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KPA
 
 | | 277 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KPK
 
 | | 309 K cryoEM structure of Sso-KARI in complex with Mg2+ | | Descriptor: | Ketol-acid reductoisomerase, MAGNESIUM ION | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KPH
 
 | | 343 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6KQO
 
 | | 328 K cryoEM structure of Sso-KARI in complex with Mg2+, NADH and CPD | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Chen, C.Y, Chang, Y.C, Lin, B.L, Huang, C.H, Tsai, M.D. | | Deposit date: | 2019-08-18 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Temperature-Resolved Cryo-EM Uncovers Structural Bases of Temperature-Dependent Enzyme Functions.
J.Am.Chem.Soc., 141, 2019
|
|
