7N6Z
 
 | | Crystal Structure of PI5P4KIIAlpha | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7N
 
 | | Crystal Structure of PI5P4KIIAlpha complex with Volasertib | | Descriptor: | N-{trans-4-[4-(cyclopropylmethyl)piperazin-1-yl]cyclohexyl}-4-{[(7R)-7-ethyl-5-methyl-8-(1-methylethyl)-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxybenzamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7O
 
 | | Crystal Structure of PI5P4KIIAlpha complex with Palbociclib | | Descriptor: | 6-ACETYL-8-CYCLOPENTYL-5-METHYL-2-[(5-PIPERAZIN-1-YLPYRIDIN-2-YL)AMINO]PYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N81
 
 | | Crystal Structure of PI5P4KIIBeta complex with CC260 | | Descriptor: | (7R)-8-cyclopentyl-7-(cyclopentylmethyl)-2-[(3,5-dichloro-4-hydroxyphenyl)amino]-5-methyl-7,8-dihydropteridin-6(5H)-one, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-11 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7J
 
 | | Crystal Structure of PI5P4KIIAlpha complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, ... | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N80
 
 | | Crystal Structure of PI5P4KIIBeta | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-11 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7M
 
 | | Crystal Structure of PI5P4KIIAlpha complex with BI-2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7K
 
 | | Crystal Structure of PI5P4KIIAlpha complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, ... | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N7L
 
 | | Crystal Structure of PI5P4KIIAlpha complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7N71
 
 | | Crystal Structure of PI5P4KIIAlpha | | Descriptor: | Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1EJC
 
 | | Crystal structure of unliganded mura (type2) | | Descriptor: | GLYCEROL, PHOSPHATE ION, UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYLTRANSFERASE | | Authors: | Eschenburg, S, Schonbrunn, E. | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative X-ray analysis of the un-liganded fosfomycin-target murA.
Proteins, 40, 2000
|
|
7D9F
 
 | | SpdH Spermidine dehydrogenase SeMet Structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9I
 
 | | SpdH Spermidine dehydrogenase D282A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, Spermidine dehydrogenase, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9H
 
 | | SpdH Spermidine dehydrogenase N33 truncation structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9G
 
 | | SpdH Spermidine dehydrogenase native structure | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, Spermidine dehydrogenase, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
7D9J
 
 | | SpdH Spermidine dehydrogenase Y443A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of Pseudomonas aeruginosa spermidine dehydrogenase: a polyamine oxidase with a novel heme-binding fold.
Febs J., 289, 2022
|
|
5Z2V
 
 | | Crystal structure of RecR from Pseudomonas aeruginosa PAO1 | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of RecR, a member of the RecFOR DNA-repair pathway, from Pseudomonas aeruginosa PAO1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1SNN
 
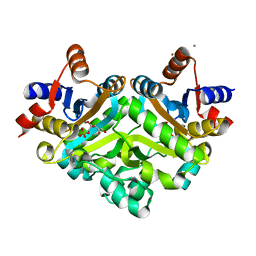 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Steinbacher, S, Schiffmann, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-03-11 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metal sites in 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with the substrate ribulose 5-phosphate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4OS7
 
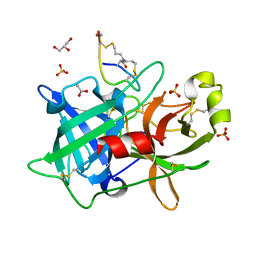 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK607 (bicyclic) | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS2
 
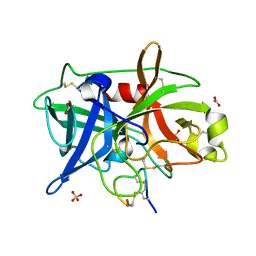 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK602 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS5
 
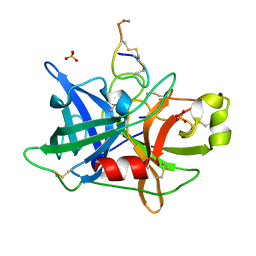 | |
4OS1
 
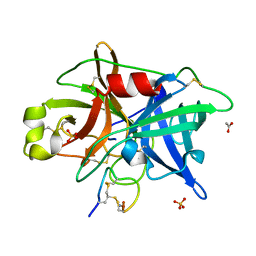 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK601 (bicyclic 1) | | Descriptor: | ACETATE ION, SULFATE ION, Urokinase-type plasminogen activator, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
4OS4
 
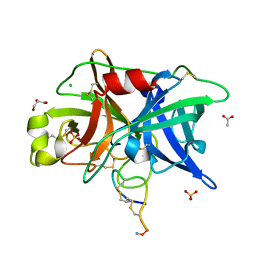 | | Crystal structure of urokinase-type plasminogen activator (uPA) complexed with bicyclic peptide UK603 (bicyclic 1) | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2014-02-12 | | Release date: | 2014-09-24 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides.
Nat Chem, 6, 2014
|
|
1S3M
 
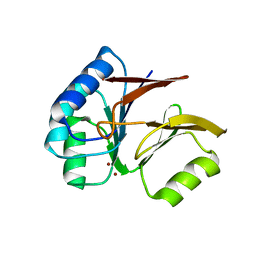 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, NICKEL (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
1S3N
 
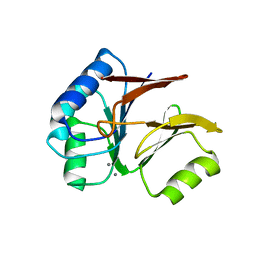 | | Structural and Functional Characterization of a Novel Archaeal Phosphodiesterase | | Descriptor: | Hypothetical protein MJ0936, MANGANESE (II) ION | | Authors: | Chen, S, Busso, D, Yakunin, A.F, Kuznetsova, E, Proudfoot, M, Jancrick, J, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of a novel phosphodiesterase from Methanococcus jannaschii
J.Biol.Chem., 279, 2004
|
|
