6NY9
 
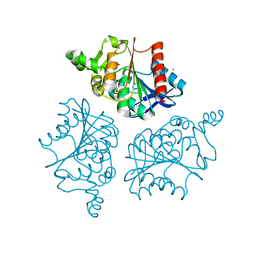 | | Alpha/beta hydrolase domain-containing protein 10 from mouse | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mycophenolic acid acyl-glucuronide esterase, mitochondrial, ... | | Authors: | Cao, Y, Rice, P.A, Dickinson, B.C. | | Deposit date: | 2019-02-11 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5.
Nat.Chem.Biol., 15, 2019
|
|
1CCJ
 
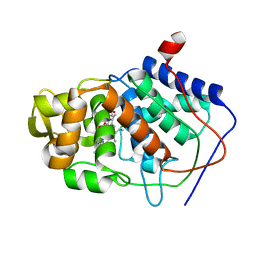 | | CONFORMER SELECTION BY LIGAND BINDING OBSERVED WITH PROTEIN CRYSTALLOGRAPHY | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein conformer selection by ligand binding observed with crystallography.
Protein Sci., 7, 1998
|
|
1CCL
 
 | |
8PHY
 
 | |
4J9U
 
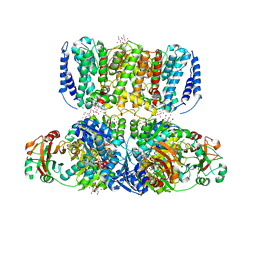 | | Crystal Structure of the TrkH/TrkA potassium transport complex | | Descriptor: | HEXATANTALUM DODECABROMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-02-17 | | Release date: | 2013-04-03 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the TrkH ion channel by its associated RCK protein TrkA.
Nature, 496, 2013
|
|
3PJZ
 
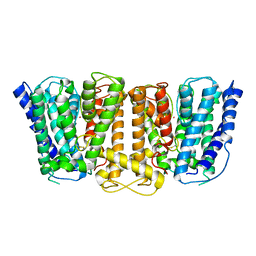 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
3QNQ
 
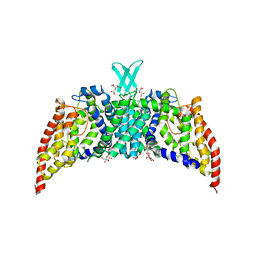 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
1A2G
 
 | |
1A2F
 
 | |
8OXL
 
 | |
8OXK
 
 | |
8OXJ
 
 | |
8OXI
 
 | |
8OXH
 
 | |
4J9V
 
 | | Crystal Structure of the TrkA Gating ring bound to ATP-gamma-S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Potassium uptake protein TrkA, ... | | Authors: | Huang, H, Levin, E.J, Jin, X, Cao, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-02-17 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Gating of the TrkH ion channel by its associated RCK protein TrkA.
Nature, 496, 2013
|
|
4EZC
 
 | | Crystal Structure of the UT-B Urea Transporter from Bos Taurus | | Descriptor: | OCTANOIC ACID (2-HYDROXY-1-HYDROXYMETHYL-HEPTADEC-3-ENYL)-AMIDE, Urea transporter 1, beta-D-glucopyranose, ... | | Authors: | Cao, Y, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2012-05-02 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure and permeation mechanism of a mammalian urea transporter.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4EZD
 
 | | Crystal Structure of the UT-B Urea Transporter from Bos Taurus Bound to Selenourea | | Descriptor: | OCTANOIC ACID (2-HYDROXY-1-HYDROXYMETHYL-HEPTADEC-3-ENYL)-AMIDE, Urea transporter 1, beta-D-glucopyranose, ... | | Authors: | Cao, Y, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2012-05-02 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and permeation mechanism of a mammalian urea transporter.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8GV3
 
 | | The cryo-EM structure of GSNOR with NYY001 | | Descriptor: | (4P)-4-{2-[4-(1H-imidazol-1-yl)phenyl]-5-[3-oxo-3-(2-oxo-1,3-thiazolidin-3-yl)propyl]-1H-pyrrol-1-yl}-3-methylbenzamide, Alcohol dehydrogenase class-3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Xia, Y, Zhang, Q, Yao, D, Zhao, S, Xie, L, Ji, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The cryo-EM structure of GSNOR with NYY001
To Be Published
|
|
8JJ6
 
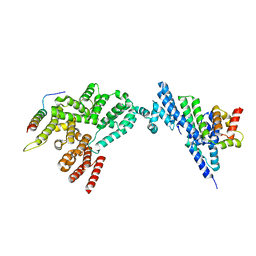 | | Structure of the NELF-BCE complex | | Descriptor: | NELF-E, Negative elongation factor B, Negative elongation factor complex member C/D | | Authors: | Wang, Z, Cao, Y, Qin, Y. | | Deposit date: | 2023-05-29 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of the human negative elongation factor NELF-B/C/E ternary complex.
Biochem.Biophys.Res.Commun., 677, 2023
|
|
7YIV
 
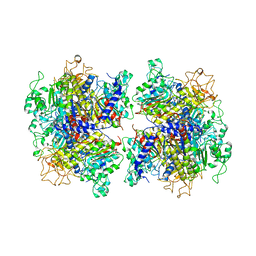 | | The Crystal Structure of Human Tissue Nonspecific Alkaline Phosphatase (ALPL) at Basic pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Cao, Y, Qin, A, Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
7Y53
 
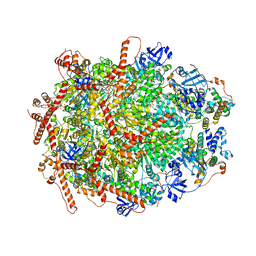 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y59
 
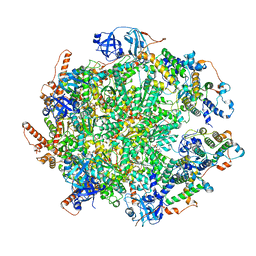 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Derlin-1, ... | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.51 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
7Y4W
 
 | | The cryo-EM structure of human ERAD retro-translocation complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Derlin-1, Transitional endoplasmic reticulum ATPase | | Authors: | Cao, Y, Rao, B, Wang, Q, Yao, D, Xia, Y, Li, W, Li, S, Shen, Y. | | Deposit date: | 2022-06-16 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | The cryo-EM structure of the human ERAD retrotranslocation complex.
Sci Adv, 9, 2023
|
|
6LUK
 
 | |
6LUJ
 
 | | Crystal structure of the SAMD1 SAM domain | | Descriptor: | Atherin, SULFATE ION | | Authors: | Cao, Y, Zhou, Y, Wang, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | The SAM domain-containing protein 1 (SAMD1) acts as a repressive chromatin regulator at unmethylated CpG islands.
Sci Adv, 7, 2021
|
|
