2IW2
 
 | | Crystal structure of human Prolidase | | Descriptor: | SODIUM ION, XAA-PRO DIPEPTIDASE | | Authors: | Mueller, U, Niesen, F.H, Roske, Y, Goetz, F, Behlke, J, Buessow, K, Heinemann, U. | | Deposit date: | 2006-06-24 | | Release date: | 2006-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of Human Prolidase: The Molecular Basis of Pd Disease
To be Published
|
|
1J75
 
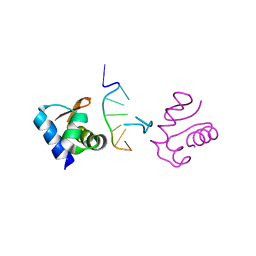 | | Crystal Structure of the DNA-Binding Domain Zalpha of DLM-1 Bound to Z-DNA | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*CP*G)-3', Tumor Stroma and Activated Macrophage Protein DLM-1 | | Authors: | Schwartz, T, Behlke, J, Lowenhaupt, K, Heinemann, U, Rich, A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-09-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins.
Nat.Struct.Biol., 8, 2001
|
|
1SAW
 
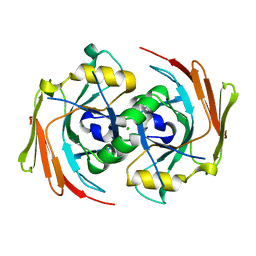 | | X-ray structure of homo sapiens protein FLJ36880 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, hypothetical protein FLJ36880 | | Authors: | Manjasetty, B.A, Niesen, F.H, Delbrueck, H, Goetz, F, Sievert, V, Buessow, K, Behlke, J, Heinemann, U. | | Deposit date: | 2004-02-09 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of fumarylacetoacetate hydrolase family member Homo sapiens FLJ36880.
Biol.Chem., 385, 2004
|
|
2OKN
 
 | | Crystal Strcture of Human Prolidase | | Descriptor: | HYDROGENPHOSPHATE ION, MANGANESE (II) ION, Xaa-Pro dipeptidase | | Authors: | Mueller, U, Niesen, F.H, Roske, Y, Goetz, F, Behlke, J, Buessow, K, Heinemann, U, Protein Structure Factory (PSF) | | Deposit date: | 2007-01-17 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Prolidase: The Molecular Basis of PD Disease.
To be Published
|
|
2YMA
 
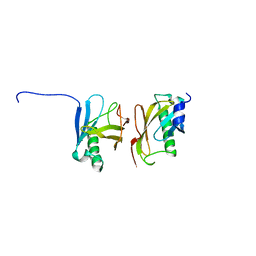 | | X-ray structure of the Yos9 dimerization domain | | Descriptor: | PROTEIN OS-9 HOMOLOG | | Authors: | Hanna, J, Schuetz, A, Zimmermann, F, Behlke, J, Sommer, T, Heinemann, U. | | Deposit date: | 2011-06-07 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Structural and Biochemical Basis of Yos9 Protein Dimerization and Possible Contribution to Self-Association of 3-Hydroxy-3-Methylglutaryl-Coenzyme a Reductase Degradation Ubiquitin-Ligase Complex.
J.Biol.Chem., 287, 2012
|
|
1U2H
 
 | | X-ray Structure of the N-terminally truncated human APEP-1 | | Descriptor: | Aortic preferentially expressed protein 1 | | Authors: | Manjasetty, B.A, Scheich, C, Roske, Y, Niesen, F.H, Gotz, F, Bussow, K, Heinemann, U. | | Deposit date: | 2004-07-19 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | X-ray structure of engineered human Aortic Preferentially Expressed Protein-1 (APEG-1)
Bmc Struct.Biol., 5, 2005
|
|
3ZJC
 
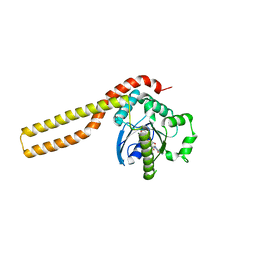 | |
3JZ7
 
 | |
3C4J
 
 | | ABC protein ArtP in complex with ATP-gamma-S | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-30 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
2JJZ
 
 | | Crystal Structure of Human Iba2, orthorhombic crystal form | | Descriptor: | ACETATE ION, CHLORIDE ION, IONIZED CALCIUM-BINDING ADAPTER MOLECULE 2, ... | | Authors: | Schulze, J.O, Quedenau, C, Roske, Y, Turnbull, A, Mueller, U, Heinemann, U, Buessow, K. | | Deposit date: | 2008-05-15 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Human Iba Proteins.
FEBS J., 275, 2008
|
|
3LJB
 
 | |
4FVF
 
 | |
4FVG
 
 | |
4FVJ
 
 | |
3C41
 
 | | ABC protein ArtP in complex with AMP-PNP/Mg2+ | | Descriptor: | Amino acid ABC transporter (ArtP), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Thaben, P.F, Eckey, V, Scheffel, F, Saenger, W, Schneider, E, Vahedi-Faridi, A. | | Deposit date: | 2008-01-29 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of the ATP-binding cassette (ABC) Protein ArtP from Geobacillus stearothermophilus in complexes with nucleotides and nucleotide analogs reveal an intermediate semiclosed dimer in the post hydrolyses state and an asymmetry in the dimerisation region
To be Published
|
|
2VTG
 
 | | Crystal Structure of Human Iba2, trigonal crystal form | | Descriptor: | ACETATE ION, IONIZED CALCIUM-BINDING ADAPTER MOLECULE 2, ZINC ION | | Authors: | Schulze, J.O, Quedenau, C, Roske, Y, Turnbull, A, Mueller, U, Heinemann, U, Buessow, K. | | Deposit date: | 2008-05-15 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and Functional Characterization of Human Iba Proteins.
FEBS J., 275, 2008
|
|
2XTO
 
 | | Crystal structure of GDP-bound human GIMAP2, amino acid residues 21- 260 | | Descriptor: | GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XTP
 
 | |
2XTN
 
 | | Crystal structure of GTP-bound human GIMAP2, amino acid residues 1- 234 | | Descriptor: | GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2XTM
 
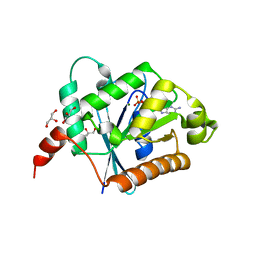 | | Crystal structure of GDP-bound human GIMAP2, amino acid residues 1- 234 | | Descriptor: | GLYCEROL, GTPASE IMAP FAMILY MEMBER 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schwefel, D, Froehlich, C, Daumke, O. | | Deposit date: | 2010-10-11 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Oligomerization in Septin-Like Gtpase of Immunity-Associated Protein 2 (Gimap2)
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1XMK
 
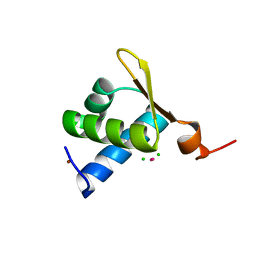 | | The Crystal structure of the Zb domain from the RNA editing enzyme ADAR1 | | Descriptor: | CADMIUM ION, CHLORIDE ION, Double-stranded RNA-specific adenosine deaminase, ... | | Authors: | Athanasiadis, A, Placido, D, Maas, S, Brown II, B.A, Lowenhaupt, K, Rich, A. | | Deposit date: | 2004-10-03 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The Crystal Structure of the Z[beta] Domain of the RNA-editing Enzyme ADAR1 Reveals Distinct Conserved Surfaces Among Z-domains.
J.Mol.Biol., 351, 2005
|
|
1SZ7
 
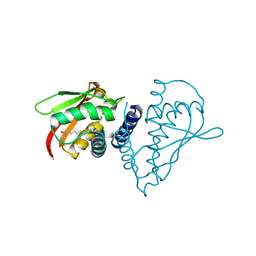 | | Crystal structure of Human Bet3 | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3 | | Authors: | Turnbull, A.P, Prinz, B, Holz, C, Behlke, J, Schultchen, J, Delbrueck, H, Niesen, F.H, Lang, C, Heinemann, U. | | Deposit date: | 2004-04-05 | | Release date: | 2005-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization
Embo J., 24, 2005
|
|
2QYP
 
 | |
2R0R
 
 | |
2R1Q
 
 | |
