3J2P
 
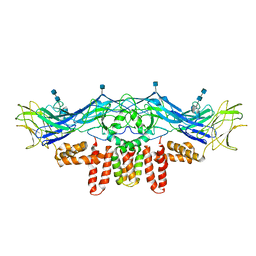 | | CryoEM structure of Dengue virus envelope protein heterotetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Zhang, X, Ge, P, Yu, X, Brannan, J.M, Bi, G, Zhang, Q, Schein, S, Zhou, Z.H. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the mature dengue virus at 3.5-A resolution.
Nat.Struct.Mol.Biol., 20, 2012
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
3J9D
 
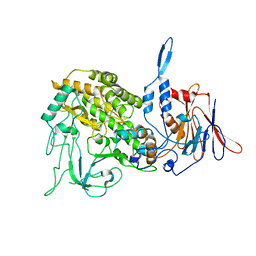 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | Outer capsid protein VP2, ZINC ION | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J9E
 
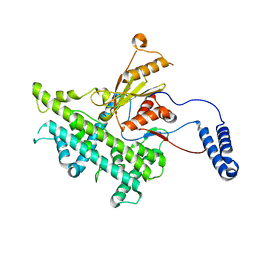 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | VP5 | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3JB7
 
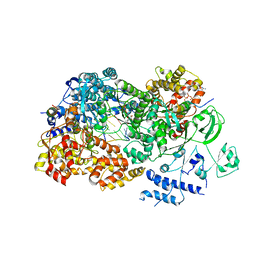 | | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus | | Descriptor: | CPV RNA-dependent RNA polymerase, CYTIDINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Ding, K, Yu, X.K, Chang, W, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-08-03 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus.
Nature, 527, 2015
|
|
1KJS
 
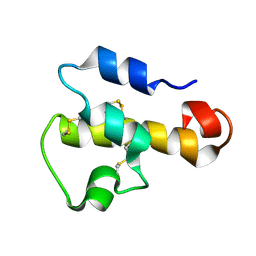 | | NMR SOLUTION STRUCTURE OF C5A AT PH 5.2, 303K, 20 STRUCTURES | | Descriptor: | C5A | | Authors: | Zhang, X, Boyar, W, Toth, M, Wennogle, L, Gonnella, N.C. | | Deposit date: | 1997-01-09 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy.
Proteins, 28, 1997
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
9NS9
 
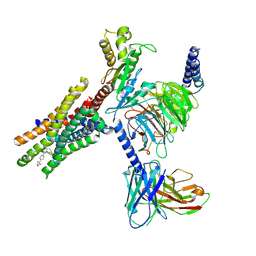 | | Cryo-EM structure of Gi-coupled FFA2 in complex with TUG-1375 and compound 187 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, 4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-7-(2-fluorobenzene-1-sulfonyl)-2-methyl-5H-pyrrolo[3,2-d]pyrimidin-6-amine, Free fatty acid receptor 2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2025-03-16 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric modulation and biased signalling at free fatty acid receptor 2.
Nature, 2025
|
|
5K3H
 
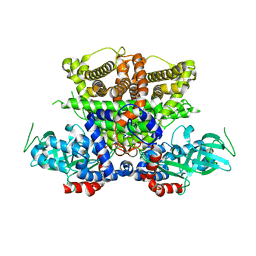 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-II | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5K3I
 
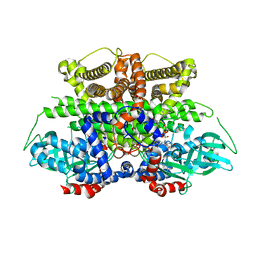 | | Crystal structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans complexed with FAD and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Acyl-coenzyme A oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1NUR
 
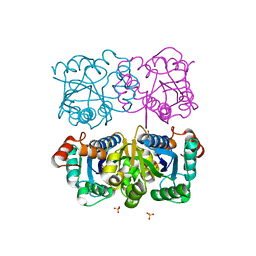 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE | | Descriptor: | FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUT
 
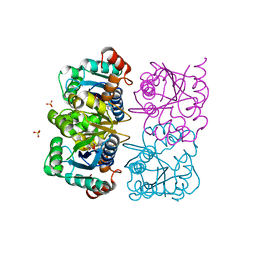 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUU
 
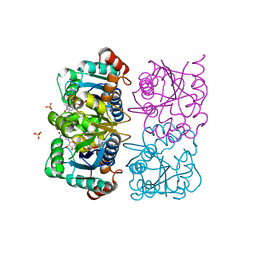 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH NAD | | Descriptor: | FKSG76, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUQ
 
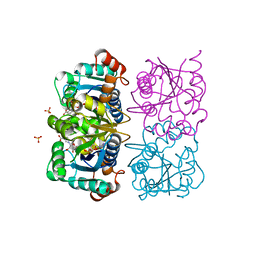 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH NaAD | | Descriptor: | FKSG76, NICOTINIC ACID ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUS
 
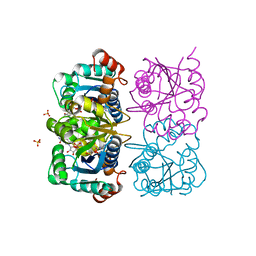 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG AND NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, ... | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1NUP
 
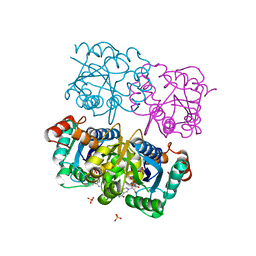 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEX WITH NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, FKSG76, SULFATE ION | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
2Y3A
 
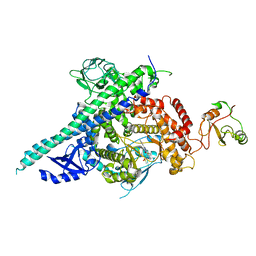 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
1NCN
 
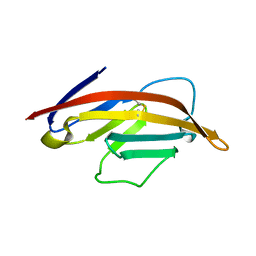 | |
3J27
 
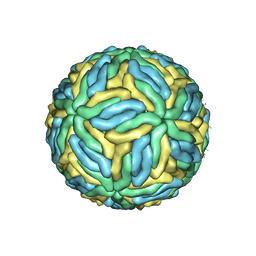 | | CryoEM structure of Dengue virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Zhang, X, Ge, P, Yu, X, Brannan, J.M, Bi, G, Zhang, Q, Schein, S, Zhou, Z.H. | | Deposit date: | 2012-09-26 | | Release date: | 2012-12-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the mature dengue virus at 3.5-A resolution.
Nat.Struct.Mol.Biol., 20, 2012
|
|
1H0C
 
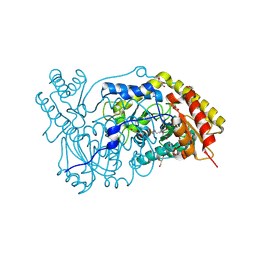 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
2CSE
 
 | | Features of Reovirus Outer-Capsid Protein mu1 Revealed by Electron and Image Reconstruction of the virion at 7.0-A Resolution | | Descriptor: | Minor core protein lambda 3, Sigma 2 protein, guanylyltransferase, ... | | Authors: | Zhang, X, Ji, Y, Zhang, L, Harrison, S.C, Marinescu, D.C, Nibert, M.L, Baker, T.S. | | Deposit date: | 2005-05-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution.
Structure, 13, 2005
|
|
4Y4N
 
 | | Thiazole synthase Thi4 from Methanococcus igneus | | Descriptor: | 2-[(E)-[(4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-ylidene]amino]ethanoic acid, FE (II) ION, Putative ribose 1,5-bisphosphate isomerase | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
4GJT
 
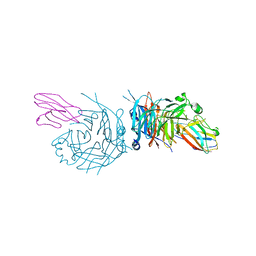 | | complex structure of nectin-4 bound to MV-H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | Authors: | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
1F3L
 
 | |
