7Y7F
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(ManQ34) and mRNA(GAC) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7E
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7G
 
 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7H
 
 | | Structure of the Bacterial Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAC) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7Y7D
 
 | | Structure of the Bacterial Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2022-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
2ZBR
 
 | | Crystal structure of ribosomal protein L11 methyltransferase from Thermus thermophilus in complex with S-adenosyl-ornithine | | Descriptor: | Ribosomal protein L11 methyltransferase, SINEFUNGIN | | Authors: | Kaminishi, T, Sakai, H, Takemoto-Hori, C, Terada, T, Nakagawa, N, Maoka, N, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-26 | | Release date: | 2008-11-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribosomal protein L11 methyltransferase from Thermus thermophilus
To be Published
|
|
7YNW
 
 | |
7YNU
 
 | |
7YNV
 
 | |
7E3D
 
 | | Crystal structure of human acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
7E3H
 
 | | Crystal structure of human acetylcholinesterase in complex with donepezil | | Descriptor: | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
6KRO
 
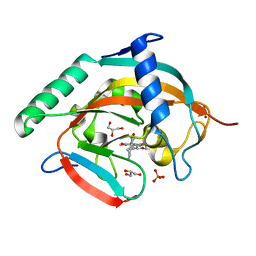 | | Tankyrase-2 in complex with RK-582 | | Descriptor: | 6-[(2S,6R)-2,6-dimethylmorpholin-4-yl]-4-fluoranyl-1-methyl-1'-(8-methyl-4-oxidanylidene-3,5,6,7-tetrahydropyrido[2,3-d]pyrimidin-2-yl)spiro[indole-3,4'-piperidine]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2019-08-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Discovery of an Orally Efficacious Spiroindolinone-Based Tankyrase Inhibitor for the Treatment of Colon Cancer.
J.Med.Chem., 63, 2020
|
|
1IUG
 
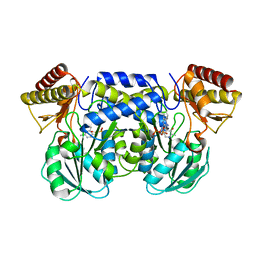 | | The crystal structure of aspartate aminotransferase which belongs to subgroup IV from Thermus thermophilus | | Descriptor: | PHOSPHATE ION, putative aspartate aminotransferase | | Authors: | Katsura, Y, Shirouzu, M, Yamaguchi, H, Ishitani, R, Nureki, O, Kuramitsu, S, Hayashi, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-04 | | Release date: | 2003-11-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative aspartate aminotransferase belonging to subgroup IV.
Proteins, 55, 2004
|
|
1IUL
 
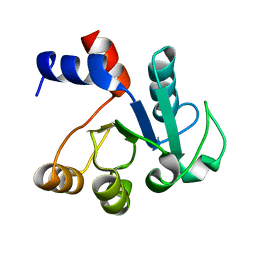 | | The structure of cell-free ID.343 from Thermus thermophilus | | Descriptor: | hypothetical protein TT1466 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a conserved CoA-binding protein synthesized by a cell-free system.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1J1V
 
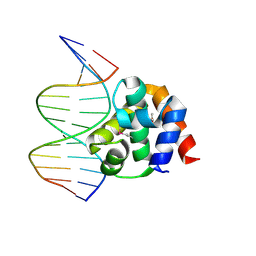 | | Crystal structure of DnaA domainIV complexed with DnaAbox DNA | | Descriptor: | 5'-D(*CP*CP*TP*GP*TP*GP*GP*AP*TP*AP*AP*CP*A)-3', 5'-D(*TP*GP*TP*TP*AP*TP*CP*CP*AP*CP*AP*GP*G)-3', Chromosomal replication initiator protein dnaA | | Authors: | Fujikawa, N, Kurumizaka, H, Nureki, O, Terada, T, Shirouzu, M, Katayama, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-12-18 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of replication origin recognition by the DnaA protein
NUCLEIC ACIDS RES., 31, 2003
|
|
5YZA
 
 | | Crystal Structure of Human CRMP-2 with S522D mutation | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Sumi, T, Imasaki, T, Aoki, M, Sakai, N, Nitta, E, Shirouzu, M, Nitta, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into the Altering Function of CRMP2 by Phosphorylation.
Cell Struct. Funct., 43, 2018
|
|
5YZ5
 
 | | Crystal Structure of Human CRMP-2 with T509D-T514D-S518D-S522D mutations | | Descriptor: | Dihydropyrimidinase-related protein 2, SULFATE ION | | Authors: | Imasaki, T, Sumi, T, Aoki, M, Sakai, N, Nitta, E, Shirouzu, M, Nitta, R. | | Deposit date: | 2017-12-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Altering Function of CRMP2 by Phosphorylation.
Cell Struct. Funct., 43, 2018
|
|
6AJ4
 
 | |
7DQC
 
 | | Crystal structure of nucleotide-free mutant A(S23C)3B(N64C)3 complex from Enterococcus hirae V-ATPase | | Descriptor: | GLYCEROL, V-type sodium ATPase catalytic subunit A, V-type sodium ATPase subunit B | | Authors: | Maruyama, S, Suzuki, K, Mizutani, K, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Murata, M. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | The combination of high-speed AFM and X-ray crystallography reveals rotary catalytic mechanism of shaftless V1-ATPase
To Be Published
|
|
5ZE9
 
 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
5B0U
 
 | |
7CLX
 
 | | Crystal structure of the DOCK8 DHR-1 domain | | Descriptor: | Dedicator of cytokinesis protein 8 | | Authors: | Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S, Fukui, Y, Uruno, T. | | Deposit date: | 2020-07-22 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A conserved PI(4,5)P2-binding domain is critical for immune regulatory function of DOCK8.
Life Sci Alliance, 4, 2021
|
|
7CLY
 
 | | Structure of the DOCK8 DHR-1 domain crystallized with di-C8-phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | Dedicator of cytokinesis protein 8 | | Authors: | Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S, Fukui, Y, Uruno, T. | | Deposit date: | 2020-07-22 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | A conserved PI(4,5)P2-binding domain is critical for immune regulatory function of DOCK8.
Life Sci Alliance, 4, 2021
|
|
1IUJ
 
 | | The structure of TT1380 protein from thermus thermophilus | | Descriptor: | ZINC ION, hypothetical protein TT1380 | | Authors: | Wada, T, Shirouzu, M, Park, S.-Y, Tame, J.R.H, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the conserved hypothetical protein TT1380 from Thermus thermophilus HB8
Proteins, 55, 2004
|
|
7CD1
 
 | | Crystal structure of inhibitory Smad, Smad7 | | Descriptor: | CHLORIDE ION, Mothers against decapentaplegic homolog 7, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for inhibitory effects of Smad7 on TGF-beta family signaling.
J.Struct.Biol., 212, 2020
|
|
