5OT4
 
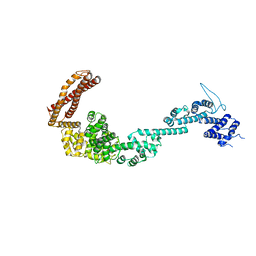 | | Structure of the Legionella pneumophila effector RidL (1-866) | | Descriptor: | GLYCEROL, Interaptin | | Authors: | Romano-Moreno, M, Rojas, A.L, Lucas, M, Isupov, M.N, Hierro, A. | | Deposit date: | 2017-08-20 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for the subversion of the retromer coat by the Legionella effector RidL.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6I50
 
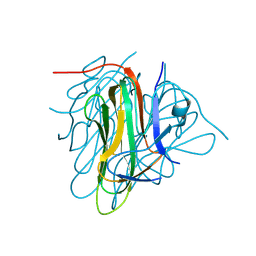 | |
8QX4
 
 | | Sulfolobus acidocaldarius Archaellum filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Flagellin, ... | | Authors: | Isupov, M.N, Gaines, M, McLaren, M, Daum, B. | | Deposit date: | 2023-10-22 | | Release date: | 2024-10-30 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.03 Å) | | Cite: | Towards a molecular picture of the archaeal cell surface.
Nat Commun, 15, 2024
|
|
5AE6
 
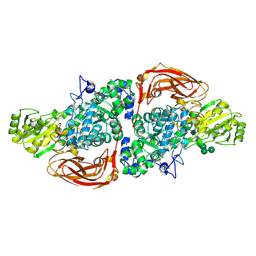 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) in complex with 4-thioxylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-XYLOSIDASE, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-08-26 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
5A7M
 
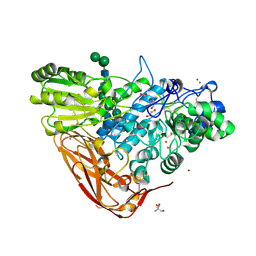 | | The structure of Hypocrea jecorina beta-xylosidase Xyl3A (Bxl1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mikkelsen, N.E, Gudmundsson, M, Karkehabadi, S, Hansson, H, Sandgren, M, Larenas, E, Mitchinson, C, Keleman, B, Kaper, T. | | Deposit date: | 2015-07-08 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Th Crystal Structure of a Fungal Glycoside Hydrolase Family 3 Beta-Xylosidase, Xyl3A from Hypocrea Jecorina
To be Published
|
|
4B5Q
 
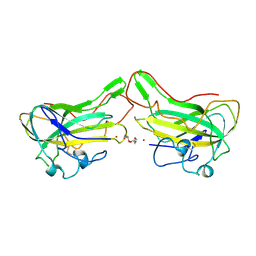 | | The lytic polysaccharide monooxygenase GH61D structure from the basidiomycota fungus Phanerochaete chrysosporium | | Descriptor: | COPPER (II) ION, GLYCEROL, GLYCOSIDE HYDROLASE FAMILY 61 PROTEIN D, ... | | Authors: | Wu, M, Beckham, G.T, Larsson, A.M, Ishida, T, Kim, S, Crowley, M.F, Payne, C.M, Horn, S.J, Westereng, B, Stahlberg, J, Eijsink, V.G.H, Sandgren, M. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Computational Characterization of the Lytic Polysaccharide Monooxygenase Gh61D from the Basidiomycota Fungus Phanerochaete Chrysosporium
J.Biol.Chem., 288, 2013
|
|
5NNS
 
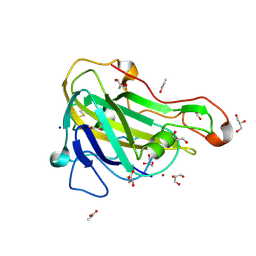 | | Crystal structure of HiLPMO9B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, COPPER (II) ION, ... | | Authors: | Dimarogona, M, Sandgren, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and molecular dynamics studies of a C1-oxidizing lytic polysaccharide monooxygenase from Heterobasidion irregulare reveal amino acids important for substrate recognition.
FEBS J., 285, 2018
|
|
6HVO
 
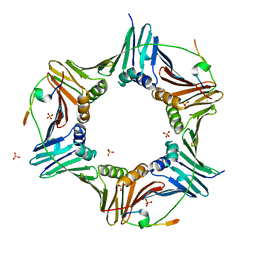 | | Crystal structure of human PCNA in complex with three peptides of p12 subunit of human polymerase delta | | Descriptor: | DNA polymerase delta subunit 4, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Gonzalez-Magana, A, Romano-Moreno, M, Rojas, A.L, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The p12 subunit of human polymerase delta uses an atypical PIP box for molecular recognition of proliferating cell nuclear antigen (PCNA).
J.Biol.Chem., 294, 2019
|
|
3NLL
 
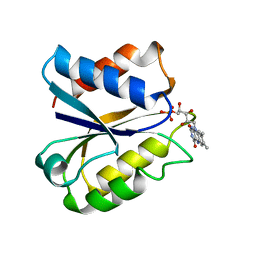 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57A OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-10 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
5NUL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN MUTANT: G57T SEMIQUINONE (150K) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-20 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
8POF
 
 | | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative glycosyl hydrolase family7, SODIUM ION | | Authors: | Haataja, T, Sandgren, M, Hansson, H, Stahlberg, J. | | Deposit date: | 2023-07-04 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites.
Febs J., 291, 2024
|
|
5NLL
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN: OXIDIZED | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1996-12-23 | | Release date: | 1997-03-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
8QSI
 
 | |
8QSY
 
 | | Portal capsid interface of full Haloferax tailed virus 1. | | Descriptor: | Capsid stabilization protein, HK97 gp5-like major capsid protein, HK97 gp6-like/SPP1 gp15-like head-tail connector, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-11 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
8QQN
 
 | |
8QPG
 
 | | Turret of Haloferax tailed virus 1 | | Descriptor: | MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ZINC ION, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
8QPQ
 
 | | C1 turret to capsid interface of full Haloferax tailed virus 1 adjacent to the portal-capsid interface. | | Descriptor: | HK97 gp5-like major capsid protein, MAGNESIUM ION, Prokaryotic polysaccharide deacetylase, ... | | Authors: | Zhang, D, Daum, B, Isupov, M.N, McLaren, M. | | Deposit date: | 2023-10-02 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structure of Haloferax tailed virus 1
To Be Published
|
|
3ZDT
 
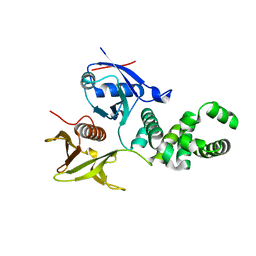 | | Crystal structure of basic patch mutant FAK FERM domain FAK31- 405 K216A, K218A, R221A, K222A | | Descriptor: | FOCAL ADHESION KINASE 1 | | Authors: | Goni, G.M, Epifano, C, Boskovic, J, Camacho-Artacho, M, Zhou, J, Martin, M.T, Eck, M.J, Kremer, L, Graeter, F, Gervasio, F.L, Perez-Moreno, M, Lietha, D. | | Deposit date: | 2012-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Phosphatidylinositol 4,5-Bisphosphate Triggers Activation of Focal Adhesion Kinase by Inducing Clustering and Conformational Changes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2RDO
 
 | | 50S subunit with EF-G(GDPNP) and RRF bound | | Descriptor: | 23S RIBOSOMAL RNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Gao, N, Zavialov, A.V, Ehrenberg, M, Frank, J. | | Deposit date: | 2007-09-24 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits.
J.Mol.Biol., 374, 2007
|
|
4CSI
 
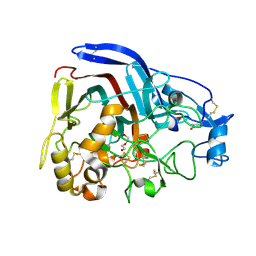 | | Crystal structure of the thermostable Cellobiohydrolase Cel7A from the fungus Humicola grisea var. thermoidea. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLULASE, DI(HYDROXYETHYL)ETHER | | Authors: | Haddad-Momeni, M, Goedegebuur, F, Hansson, H, Karkehabadi, S, Askarieh, G, Mitchinson, C, Larenas, E, Stahlberg, J, Sandgren, M. | | Deposit date: | 2014-03-07 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Crystal Structure and Cellulase Activity of the Thermostable Cellobiohydrolase Cel7A from the Fungus Humicola Grisea Var. Thermoidea.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PI8
 
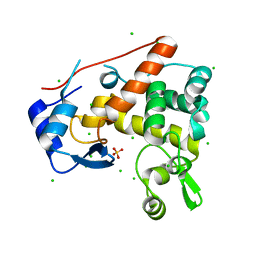 | | Crystal structure of catalytic mutant E138A of S. Aureus Autolysin E in complex with disaccharide NAG-NAM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid, Autolysin E, CHLORIDE ION, ... | | Authors: | Mihelic, M, Renko, M, Jakas, A, Turk, D. | | Deposit date: | 2014-05-08 | | Release date: | 2015-10-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | The mechanism behind the selection of two different cleavage sites in NAG-NAM polymers
Iucrj, 4, 2017
|
|
1NK3
 
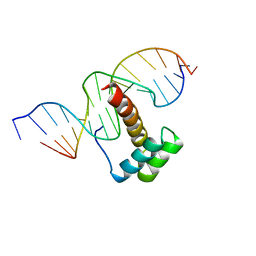 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
1NK2
 
 | | VND/NK-2 HOMEODOMAIN/DNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*CP*CP*AP*CP*TP*TP*GP*AP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*TP*CP*AP*AP*GP*TP*GP*GP*CP*TP*GP*T)-3'), HOMEOBOX PROTEIN VND | | Authors: | Gruschus, J.M, Tsao, D.H.H, Wang, L.-H, Nirenberg, M, Ferretti, J.A. | | Deposit date: | 1998-05-06 | | Release date: | 1999-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity.
Biochemistry, 36, 1997
|
|
5AJH
 
 | | Crystal structure of Fusarium oxysporum cutinase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CUTINASE | | Authors: | Dimarogona, M, Nikolaivits, E, Kanelli, M, Christakopoulos, P, Sandgren, M, Topakas, E. | | Deposit date: | 2015-02-24 | | Release date: | 2015-09-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Studies of a Fusarium Oxysporum Cutinase with Polyethylene Terephthalate Modification Potential.
Biochim.Biophys.Acta, 1850, 2015
|
|
5OA5
 
 | | CELLOBIOHYDROLASE I (CEL7A) FROM HYPOCREA JECORINA WITH IMPROVED THERMAL STABILITY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exoglucanase 1, GLYCEROL | | Authors: | Goedegebuur, F, Hansson, H, Karkehabadi, S, Mikkelsen, N, Stahlberg, J, Sandgren, M. | | Deposit date: | 2017-06-20 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving the thermal stability of cellobiohydrolase Cel7A from Hypocrea jecorina by directed evolution.
J. Biol. Chem., 292, 2017
|
|
