6TVO
 
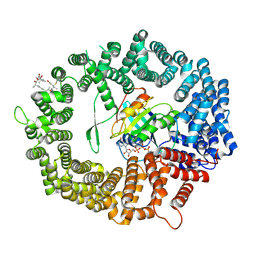 | | Human CRM1-RanGTP in complex with Leptomycin B | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-01-10 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Characterization of Inhibition Reveals Distinctive Properties for Human andSaccharomyces cerevisiaeCRM1.
J.Med.Chem., 63, 2020
|
|
2KB6
 
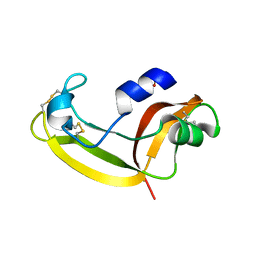 | | Solution structure of onconase C87A/C104A | | Descriptor: | Protein P-30 | | Authors: | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
6ZM2
 
 | |
5ORI
 
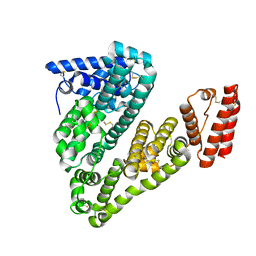 | | Structure of caprine serum albumin in orthorhombic crystal system | | Descriptor: | Albumin | | Authors: | Bujacz, A, Talaj, J.A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OTB
 
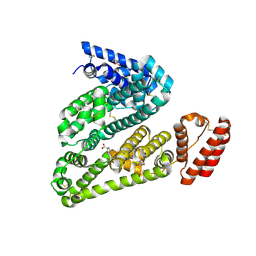 | | Structure of caprine serum albumin in P1 space group | | Descriptor: | Albumin, DI(HYDROXYETHYL)ETHER, PROLINE, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5ORF
 
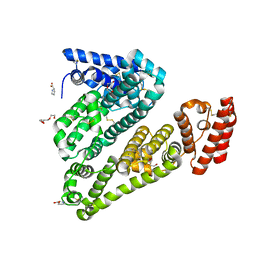 | | Structure of ovine serum albumin in P1 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, PROLINE, Serum albumin, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5OSW
 
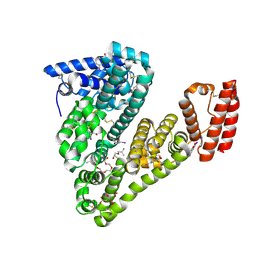 | | Structure of caprine serum albumin in complex with 3,5-diiodosalicylic acid | | Descriptor: | 2-HYDROXY-3,5-DIIODO-BENZOIC ACID, 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, Albumin, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-08-18 | | Release date: | 2017-11-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2K9I
 
 | |
2L1L
 
 | |
3NOK
 
 | | Crystal structure of Myxococcus xanthus Glutaminyl Cyclase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
3NOL
 
 | | Crystal structure of Zymomonas mobilis Glutaminyl Cyclase (trigonal form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
6SUR
 
 | | The Rab33B-Atg16L1 crystal structure | | Descriptor: | Autophagy-related protein 16-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Metje-Sprink, J, Kuehnel, K. | | Deposit date: | 2019-09-16 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.467 Å) | | Cite: | Crystal structure of the Rab33B/Atg16L1 effector complex.
Sci Rep, 10, 2020
|
|
3CBG
 
 | | Functional and Structural Characterization of a Cationdependent O-Methyltransferase from the Cyanobacterium Synechocystis Sp. Strain PCC 6803 | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)prop-2-enoic acid, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, MAGNESIUM ION, ... | | Authors: | Kopycki, J.G, Neumann, P, Stubbs, M.T. | | Deposit date: | 2008-02-22 | | Release date: | 2008-06-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and Structural Characterization of a Cation-dependent O-Methyltransferase from the Cyanobacterium Synechocystis sp. Strain PCC 6803
J.Biol.Chem., 283, 2008
|
|
3NOM
 
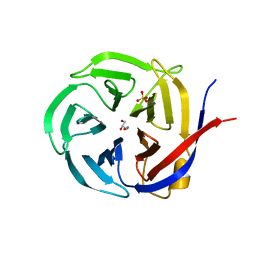 | | Crystal Structure of Zymomonas mobilis Glutaminyl Cyclase (monoclinic form) | | Descriptor: | CALCIUM ION, GLYCEROL, Glutamine cyclotransferase, ... | | Authors: | Parthier, C, Carrillo, D.R, Stubbs, M.T. | | Deposit date: | 2010-06-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural characterization of bacterial glutaminyl cyclases from Zymomonas mobilis and Myxococcus xanthus
Biol.Chem., 391, 2010
|
|
