4FA4
 
 | |
4FA5
 
 | |
4FA9
 
 | |
4FB1
 
 | |
4FAV
 
 | |
6MAE
 
 | |
6PEU
 
 | |
6PE3
 
 | |
7JII
 
 | | HRAS A59E GDP | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIF
 
 | | HRAS A59T GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIH
 
 | | HRAS A59E GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIG
 
 | | HRAS A59T GppNHp crystal 2 | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
1TMB
 
 | | MOLECULAR BASIS FOR THE INHIBITION OF HUMAN ALPHA-THROMBIN BY THE MACROCYCLIC PEPTIDE CYCLOTHEONAMIDE A | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | Authors: | Qiu, X, Padmanabhan, K.P, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1993-05-27 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of human alpha-thrombin by the macrocyclic peptide cyclotheonamide A.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XCR
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD0
 
 | | cryo-EM structure of H2BK34ub nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
 | | cryo-EM structure of unmodified nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
9CDU
 
 | | Crystal Structure of Rhombotarget A-peptide Complex | | Descriptor: | RBB_D10 peptide, Rhombotarget A, TETRAETHYLENE GLYCOL | | Authors: | Bera, A.K, Rettie, S, Kang, A, Bhardwaj, G. | | Deposit date: | 2024-06-25 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Accurate de novo design of high-affinity protein-binding macrocycles using deep learning.
Nat.Chem.Biol., 2025
|
|
9CDV
 
 | | Crystal Structure of Rhombotarget A | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Rhombotarget A | | Authors: | Bera, A.K, Rettie, S, Kang, A, Bhardwaj, G. | | Deposit date: | 2024-06-25 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accurate de novo design of high-affinity protein-binding macrocycles using deep learning.
Nat.Chem.Biol., 2025
|
|
9CDT
 
 | |
8GTH
 
 | |
3IE9
 
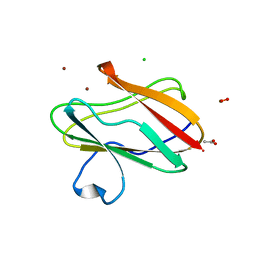 | | Structure of oxidized M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
3IEA
 
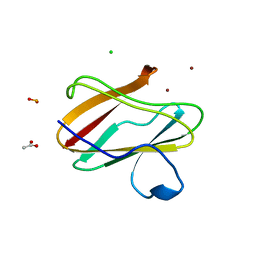 | | Structure of reduced M98L mutant of amicyanin | | Descriptor: | ACETATE ION, Amicyanin, CHLORIDE ION, ... | | Authors: | Sukumar, N, Davidson, V.L. | | Deposit date: | 2009-07-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Defining the role of the axial ligand of the type 1 copper site in amicyanin by replacement of methionine with leucine.
Biochemistry, 48, 2009
|
|
7X6S
 
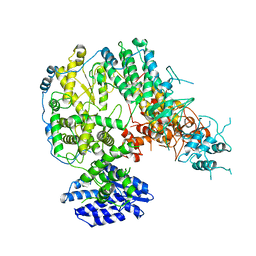 | |
7X6V
 
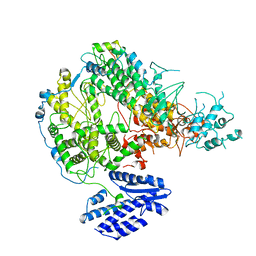 | |
