3LWA
 
 | | The Crystal Structure of a Secreted Thiol-disulfide Isomerase from Corynebacterium glutamicum to 1.75A | | Descriptor: | CALCIUM ION, Secreted thiol-disulfide isomerase | | Authors: | Stein, A.J, Weger, A, Hendricks, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Crystal Structure of a Secreted Thiol-disulfide Isomerase from Corynebacterium glutamicum to 1.75A
To be Published
|
|
3M1A
 
 | | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A | | Descriptor: | ACETATE ION, Putative dehydrogenase, SODIUM ION | | Authors: | Stein, A.J, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Short-chain Dehydrogenase from Streptomyces avermitilis to 2A
To be Published
|
|
2RIR
 
 | | Crystal structure of dipicolinate synthase, A chain, from Bacillus subtilis | | Descriptor: | CHLORIDE ION, Dipicolinate synthase, A chain, ... | | Authors: | Osipiuk, J, Quartey, P, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-12 | | Release date: | 2007-10-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of dipicolinate synthase, A chain, from Bacillus subtilis.
To be Published
|
|
2R5F
 
 | | Putative sugar-binding domain of transcriptional regulator DeoR from Pseudomonas syringae pv. tomato | | Descriptor: | SULFATE ION, Transcriptional regulator, putative | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-03 | | Release date: | 2007-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Putative sugar-binding domain of trancsriptional regulator DeoR from Pseudomonas syringae pv. tomato.
TO BE PUBLISHED
|
|
2R5S
 
 | | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Wu, R, Abdullah, J, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The crystal structure of a domain of protein VP0806 (unknown function) from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
2R9I
 
 | |
5IX8
 
 | | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822 | | Descriptor: | 1,2-ETHANEDIOL, Putative sugar ABC transport system, substrate-binding protein, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-03-23 | | Release date: | 2016-04-06 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of sugar ABC transport system, substrate-binding protein from Bordetella parapertussis 12822
To Be Published
|
|
2R8R
 
 | | Crystal structure of the N-terminal region (19..243) of sensor protein KdpD from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Nocek, B, Mulligan, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-11 | | Release date: | 2007-09-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the N-terminal region (19..243) of sensor protein KdpD from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
2RK5
 
 | |
6EX7
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase in complex with Cd ions and a hydrolyzed beta-lactam ligand - new refinement | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, CADMIUM ION, ... | | Authors: | Kim, Y, Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A. | | Deposit date: | 2017-11-07 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
6W08
 
 | | Crystal Structure of Motility Associated Killing Factor E from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Jedrzejczak, R, Joachimiak, G, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
5KZM
 
 | | Crystal structure of Tryptophan synthase alpha-beta chain complex from Francisella tularensis | | Descriptor: | ACETATE ION, CALCIUM ION, Tryptophan synthase alpha chain, ... | | Authors: | Chang, C, Michalska, K, Joachimiak, G, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-25 | | Release date: | 2016-08-10 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Conservation of the structure and function of bacterial tryptophan synthases.
Iucrj, 6, 2019
|
|
7UV5
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S/D286N mutant, in complex with a Lys48-linked di-ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, Papain-like protease nsp3, Ubiquitin, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lanham, B.T, Wydorski, P, Fushman, D, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-04-29 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.
Nat Commun, 14, 2023
|
|
7JIT
 
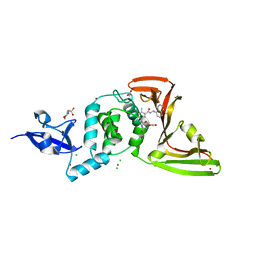 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder495 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIR
 
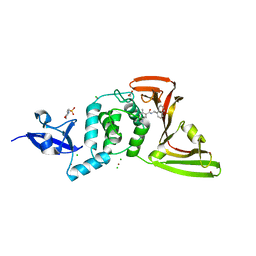 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIW
 
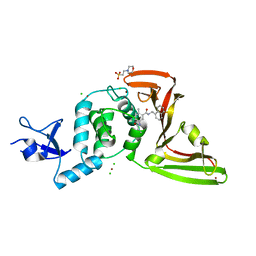 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JFQ
 
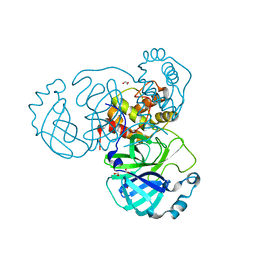 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7THH
 
 | | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Jedrzejczak, R, Endres, M, Wydorski, P, Joachimiak, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-11 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | SUD-C and Ubl2 domains of SARS CoV-2 Nsp3 protein
to be published
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | Authors: | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-25 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
6DFP
 
 | | Crystal Structure of a Tripartite Toxin Component VCA0883 from Vibrio cholerae | | Descriptor: | VCA0883 | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2022-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
4XED
 
 | | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Peptidase M14, ... | | Authors: | Michalska, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-05-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20
To Be Published
|
|
4XXT
 
 | | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from Clostridium acetobutylicum ATCC 824 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fusion of predicted Zn-dependent amidase/peptidase (Cell wall hydrolase/DD-carboxypeptidase family) and uncharacterized domain of ErfK family peptodoglycan-binding domain, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from from Clostridium acetobutylicum ATCC 824
To Be Published
|
|
4YE5
 
 | | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan synthetase penicillin-binding protein 3 | | Authors: | Cuff, M, Tan, K, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-23 | | Release date: | 2015-03-18 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | The crystal structure of a peptidoglycan synthetase from Bifidobacterium adolescentis ATCC 15703
To Be Published
|
|
5KIN
 
 | | Crystal structure of tryptophan synthase alpha beta complex from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, Tryptophan synthase alpha chain, Tryptophan synthase beta chain | | Authors: | Chang, C, Michalska, K, Bigelow, L, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-16 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conservation of the structure and function of bacterial tryptophan synthases.
Iucrj, 6, 2019
|
|
