3TX1
 
 | | X-ray crystal structure of Listeria monocytogenes EGD-e UDP-N-acetylenolpyruvylglucosamine reductase (MurB) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | X-ray crystal structure of Listeria monocytogenes EGD-e UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
TO BE PUBLISHED
|
|
3SC6
 
 | | 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, dTDP-4-dehydrorhamnose reductase | | Authors: | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-07 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose reductase (RfbD).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3SD7
 
 | | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative phosphatase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile.
TO BE PUBLISHED
|
|
3SEF
 
 | | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Shikimate 5-dehydrogenase | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH
To be Published
|
|
3SLH
 
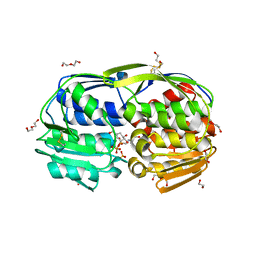 | | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 1,2-ETHANEDIOL, 3-phosphoshikimate 1-carboxyvinyltransferase, ... | | Authors: | Light, S.H, Minasov, G, Filippova, E.V, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase(AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate
To be Published
|
|
3SRT
 
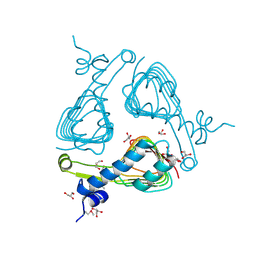 | | The crystal structure of a maltose O-acetyltransferase from Clostridium difficile 630 | | Descriptor: | GLYCEROL, Maltose O-acetyltransferase | | Authors: | Tan, K, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | The crystal structure of a maltose O-acetyltransferase from Clostridium difficile 630
To be Published
|
|
5KCK
 
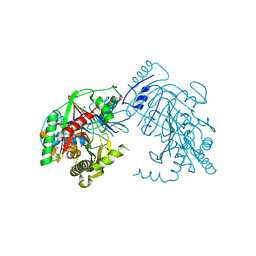 | | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4 | | Descriptor: | Anthranilate synthase component I, GLYCEROL | | Authors: | Chang, C, Michalska, K, Bigelow, L, Jedrzejczak, R, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-06 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of anthranilate synthase component I from Streptococcus pneumoniae TIGR4
To Be Published
|
|
5JQW
 
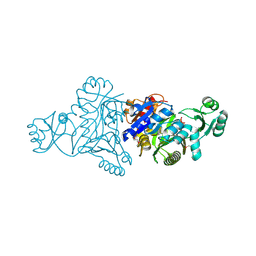 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, N5-carboxyaminoimidazole ribonucleotide synthase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-05 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
5JPI
 
 | | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD.
To Be Published
|
|
5K99
 
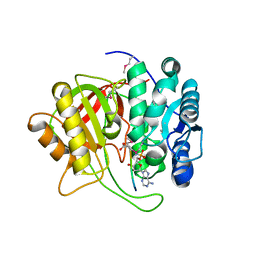 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin C, Microcin C7 self-immunity protein mccF | | Authors: | Nocek, B, Kulikovsky, A, Severinov, K, Dubiley, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-31 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC
To Be Published
|
|
3SZ3
 
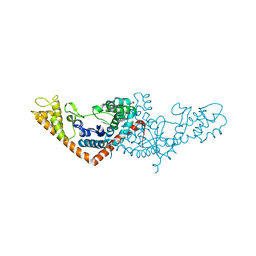 | | Crystal structure of Tryptophanyl-tRNA synthetase from Vibrio cholerae with an endogenous tryptophan | | Descriptor: | GLYCEROL, TRYPTOPHAN, Tryptophanyl-tRNA synthetase | | Authors: | Cooper, D.R, Kudritska, M, Chruszcz, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-18 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Vibrio cholerae with an endogenous tryptophan
To be Published
|
|
3SR3
 
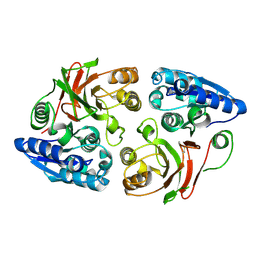 | | Crystal structure of the w180a mutant of microcin immunity protein mccf from Bacillus anthracis shows the active site loop in the open conformation. | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-06 | | Release date: | 2011-08-10 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
5JXW
 
 | | 2.25 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Neplanocin-A and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-13 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Neplanocin-A and NAD
To Be Published
|
|
3SY3
 
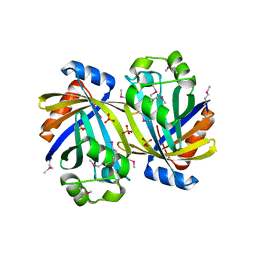 | | GBAA_1210 protein, a putative adenylate cyclase, from Bacillus anthracis | | Descriptor: | GBAA_1210 protein, PHOSPHATE ION | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-15 | | Release date: | 2011-07-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | GBAA_1210 protein, a putative adenylate cyclase, from Bacillus anthracis.
To be Published
|
|
5KBP
 
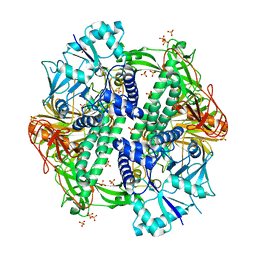 | | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583 | | Descriptor: | Glycosyl hydrolase, family 38, SULFATE ION | | Authors: | Tan, K, Chhor, G, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of an alpha-mannosidase from Enterococcus faecalis V583
To Be Published
|
|
3S42
 
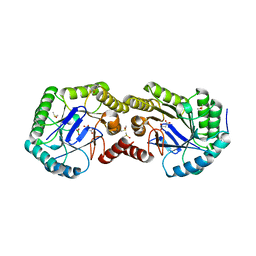 | | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site | | Descriptor: | 3-dehydroquinate dehydratase, BORIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Halavaty, A.S, Krishna, S.N, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site
To be Published
|
|
3T4X
 
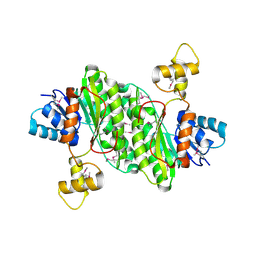 | | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Oxidoreductase, short chain dehydrogenase/reductase family | | Authors: | Filippova, E.V, Wawrzak, Z, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Short chain dehydrogenase/reductase family oxidoreductase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3T5M
 
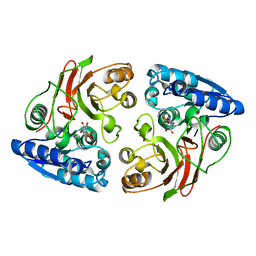 | | Crystal structure of the S112A mutant of mycrocine immunity protein (MccF) with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-27 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
5KJP
 
 | |
3T7Y
 
 | | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis | | Descriptor: | CHLORIDE ION, FORMIC ACID, SODIUM ION, ... | | Authors: | Singer, A.U, Wawrzak, Z, Skarina, T, Saikali, P, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis
TO BE PUBLISHED
|
|
3T41
 
 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | Authors: | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3T4E
 
 | | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Quinate/shikimate dehydrogenase | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD.
TO BE PUBLISHED
|
|
3S40
 
 | | The crystal structure of a diacylglycerol kinases from Bacillus anthracis str. Sterne | | Descriptor: | diacylglycerol kinase | | Authors: | Tan, K, Zhang, R, Xu, X, Cui, H, Peterson, S, Savchenko, A, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-18 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a diacylglycerol kinases from Bacillus anthracis str. Sterne
To be Published
|
|
5KKO
 
 | | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein | | Descriptor: | Uncharacterised protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-22 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein
To Be Published
|
|
5KP2
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae cocrystallized with octanoyl-CoA: hydrolzed ligand | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2, COENZYME A, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Hou, J, Zheng, H, Grabowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: hydrolzed ligand
To Be Published
|
|
