1K0N
 
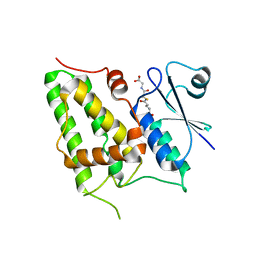 | | Chloride Intracellular Channel 1 (CLIC1) complexed with glutathione | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1, GLUTATHIONE | | Authors: | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
1K0M
 
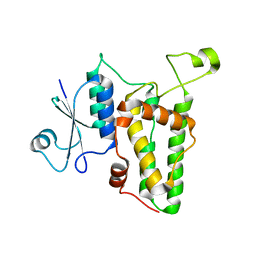 | | Crystal structure of a soluble monomeric form of CLIC1 at 1.4 angstroms | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | Authors: | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
1OMT
 
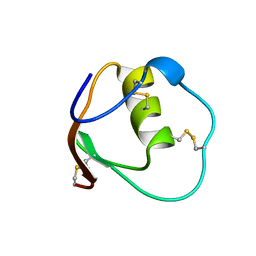 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (STANDARD NOESY ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
7Q04
 
 | | Crystal structure of TPADO in a substrate-free state | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6BI6
 
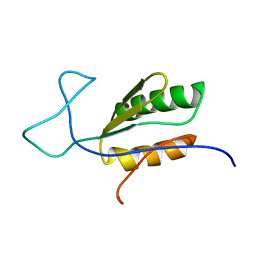 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
7Q1R
 
 | | A de novo designed homo-dimeric antiparallel coiled coil apCC-Di | | Descriptor: | ETHANOL, SODIUM ION, apCC-Di | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7Q1S
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB_var | | Descriptor: | 1,2-ETHANEDIOL, apCC-Di-A_var, apCC-Di-B_var | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7Q1T
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB | | Descriptor: | N-PROPANOL, SULFATE ION, apCC-Di-A, ... | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7Q1Q
 
 | | De novo designed homo-dimeric antiparallel helices Homomer-S | | Descriptor: | ACETATE ION, Homomer-S | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
1P9P
 
 | | The Crystal Structure of a M1G37 tRNA Methyltransferase, TrmD | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Watts, J.M, Zalacain, M, Van Thiel, A, Vitaszka, P.R, Redlak, M, Andraos-Selim, C, Rastinejad, F, Holmes, W.M. | | Deposit date: | 2003-05-12 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into Catalysis by a Knotted TrmD tRNA Methyltransferase.
J.Mol.Biol., 333, 2003
|
|
7QU1
 
 | | Machupo virus GP1 glycoprotein in complex with Fab fragment of antibody MAC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab MAC1 heavy chain, Fab MAC1 light chain, ... | | Authors: | Ng, W.M, Sahin, M, Krumm, S.A, Seow, J, Zeltina, A, Harlos, K, Paesen, G, Pinschewer, D.D, Doores, K.J, Bowden, T.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Contrasting Modes of New World Arenavirus Neutralization by Immunization-Elicited Monoclonal Antibodies.
Mbio, 13, 2022
|
|
7QU2
 
 | | Junin virus GP1 glycoprotein in complex with Fab fragment of antibody JUN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab JUN1 heavy chain, ... | | Authors: | Ng, W.M, Sahin, M, Krumm, S.A, Seow, J, Zeltina, A, Harlos, K, Paesen, G, Pinschewer, D.D, Doores, K.J, Bowden, T.A. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contrasting Modes of New World Arenavirus Neutralization by Immunization-Elicited Monoclonal Antibodies.
Mbio, 13, 2022
|
|
6EIK
 
 | | A de novo designed heptameric coiled coil CC-Hept-I24E | | Descriptor: | CC-Hept-I24E, FORMIC ACID, GLYCEROL | | Authors: | Rhys, G.G, Burton, A.J, Dawson, W.M, Thomas, F, Woolfson, D.N. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | De Novo-Designed alpha-Helical Barrels as Receptors for Small Molecules.
ACS Synth Biol, 7, 2018
|
|
7R9B
 
 | |
7R7M
 
 | |
6DHB
 
 | | Crystal structure of the human TIM-3 with bound Calcium | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | Authors: | Gandhi, A.K, Kim, W.M, Huang, Y.H, Bonsor, D, Sundberg, E, Sun, Z.-Y, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2018-05-19 | | Release date: | 2018-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution X-ray and NMR structural study of human T-cell immunoglobulin and mucin domain containing protein-3.
Sci Rep, 8, 2018
|
|
7Q06
 
 | | Crystal structure of TPADO in complex with 2-OH-TPA | | Descriptor: | 2-Hydroxyterephthalic acid, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q05
 
 | | Crystal structure of TPADO in complex with TPA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6F7J
 
 | |
7RCQ
 
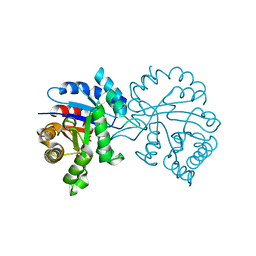 | |
7RGC
 
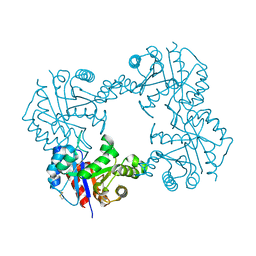 | |
6EIZ
 
 | | A de novo designed hexameric coiled coil CC-Hex2 with farnesol bound in the channel. | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, CC-Hex2 | | Authors: | Rhys, G.G, Burton, A.J, Dawson, W.M, Thomas, F, Woolfson, D.N. | | Deposit date: | 2017-09-19 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De Novo-Designed alpha-Helical Barrels as Receptors for Small Molecules.
ACS Synth Biol, 7, 2018
|
|
1ROC
 
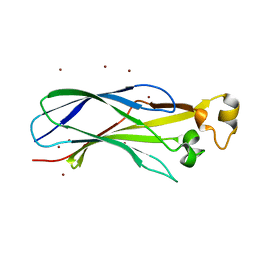 | | Crystal structure of the histone deposition protein Asf1 | | Descriptor: | Anti-silencing protein 1, BROMIDE ION | | Authors: | Daganzo, S.M, Erzberger, J.P, Lam, W.M, Skordalakes, E, Zhang, R, Franco, A.A, Brill, S.J, Adams, P.D, Berger, J.M, Kaufman, P.D. | | Deposit date: | 2003-12-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the conserved core of histone deposition protein Asf1.
Curr.Biol., 13, 2003
|
|
7QWC
 
 | |
1RGQ
 
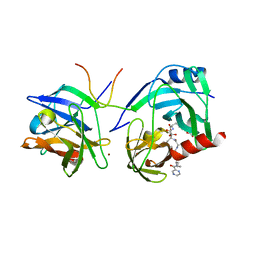 | | M9A HCV Protease complex with pentapeptide keto-amide inhibitor | | Descriptor: | N-(PYRAZIN-2-YLCARBONYL)LEUCYLISOLEUCYL-N~1~-{1-[2-({1-CARBOXY-2-[4-(PHOSPHONOOXY)PHENYL]ETHYL}AMINO)-1,1-DIHYDROXY-2-OXOETHYL]BUT-3-ENYL}-3-CYCLOHEXYLALANINAMIDE, NS3 Protease, NS4A peptide, ... | | Authors: | Liu, Y, Stoll, V.S, Richardson, P.L, Saldivar, A, Klaus, J.L, Molla, A, Kohlbrenner, W, Kati, W.M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Arch.Biochem.Biophys., 421, 2004
|
|
