2IXJ
 
 | | RmlC P aeruginosa native | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXI
 
 | | RmlC P aeruginosa with dTDP-xylose | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, S,R MESO-TARTARIC ACID, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-08 | | Release date: | 2006-07-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rmlc, a C3' and C5' Carbohydrate Epimerase, Appears to Operate Via an Intermediate with an Unusual Twist Boat Conformation.
J.Mol.Biol., 365, 2007
|
|
2IXK
 
 | |
3OHL
 
 | |
3OHO
 
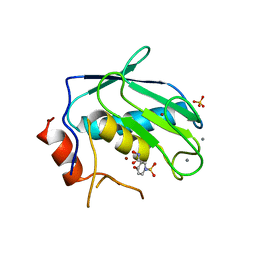 | |
1KD0
 
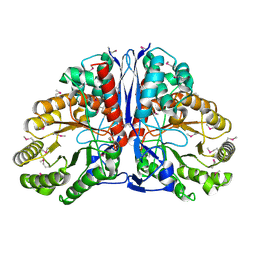 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
1KCZ
 
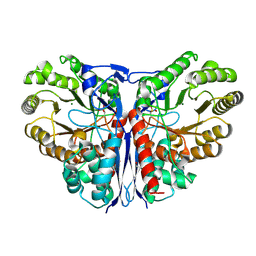 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
5FIU
 
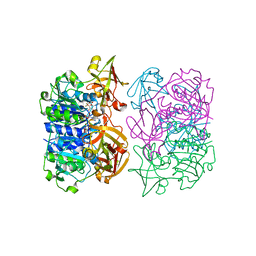 | | Binding and structural studies of a 5,5-difluoromethyl adenosine nucleoside with the fluorinase enzyme | | Descriptor: | 5'-FLUORO-5'-DEOXY-ADENOSINE SYNTHASE, 5,5-DIFLUOROMETHYL ADENOSINE, L(+)-TARTARIC ACID | | Authors: | Thompson, S, McMahon, S.A, Naismith, J.H, O'Hagan, D. | | Deposit date: | 2015-10-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Exploration of a Potential Difluoromethyl-Nucleoside Substrate with the Fluorinase Enzyme.
Bioorg.Chem., 64, 2015
|
|
1WA3
 
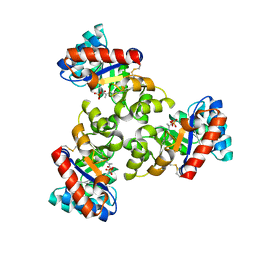 | | Mechanism of the Class I KDPG aldolase | | Descriptor: | 2-KETO-3-DEOXY-6-PHOSPHOGLUCONATE ALDOLASE, PYRUVIC ACID, SULFATE ION | | Authors: | Fullerton, S.W.B, Griffiths, J.S, Merkel, A.B, Wymer, N.J, Hutchins, M.J, Fierke, C.A, Toone, E.J, Naismith, J.H. | | Deposit date: | 2004-10-22 | | Release date: | 2005-01-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of the Class I Kdpg Aldolase.
Bioorg.Med.Chem., 14, 2006
|
|
1WAU
 
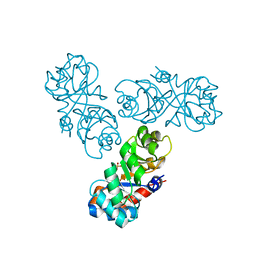 | |
1WAM
 
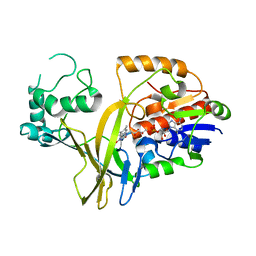 | |
1WBH
 
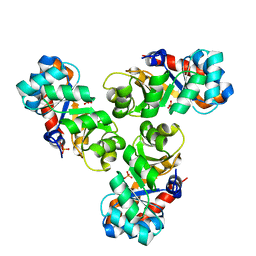 | |
9EXO
 
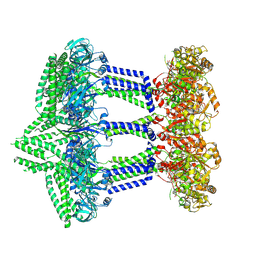 | | Wzc-K540M-2YE MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Liu, J.W, Yang, Y, Naismith, J.H. | | Deposit date: | 2024-04-08 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the phosphorylation of bacterial tyrosine kinase Wzc.
Nat Commun, 16, 2025
|
|
9EXR
 
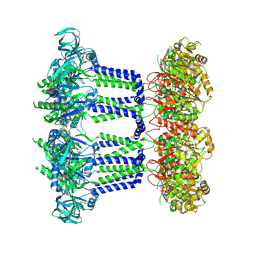 | | Wzc-K540M-3YE-N711Y MgADP C8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Liu, J.W, Yang, Y, Naismith, J.H. | | Deposit date: | 2024-04-08 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Molecular basis for the phosphorylation of bacterial tyrosine kinase Wzc.
Nat Commun, 16, 2025
|
|
9EXP
 
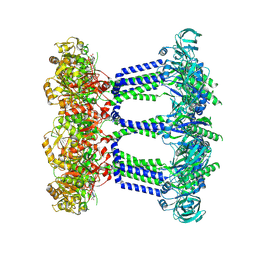 | | Wzc-K540M-2YE MgADP C8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Liu, J.W, Yang, Y, Naismith, J.H. | | Deposit date: | 2024-04-08 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for the phosphorylation of bacterial tyrosine kinase Wzc.
Nat Commun, 16, 2025
|
|
9EXQ
 
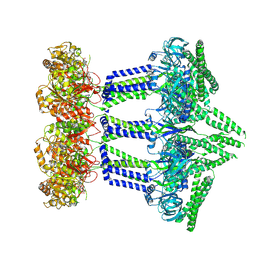 | | Wzc-K540M-3YE-N711Y MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Liu, J.W, Yang, Y, Naismith, J.H. | | Deposit date: | 2024-04-08 | | Release date: | 2025-04-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Molecular basis for the phosphorylation of bacterial tyrosine kinase Wzc.
Nat Commun, 16, 2025
|
|
2APG
 
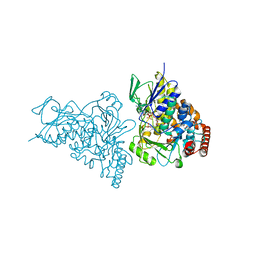 | | The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-16 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
9I2Q
 
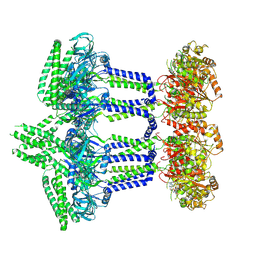 | | Wzc-K540M-3YE MgADP C1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Liu, J.W, Yang, Y, Naismith, J.H. | | Deposit date: | 2025-01-22 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for the phosphorylation of bacterial tyrosine kinase Wzc.
Nat Commun, 16, 2025
|
|
2IYD
 
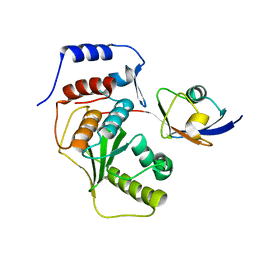 | | SENP1 covalent complex with SUMO-2 | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1, SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2IYC
 
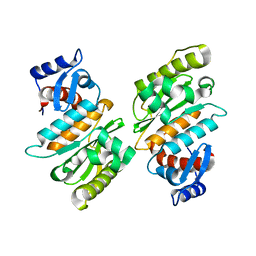 | | SENP1 native structure | | Descriptor: | SENTRIN-SPECIFIC PROTEASE 1 | | Authors: | Dong, C, Naismith, J.H. | | Deposit date: | 2006-07-14 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Senp1 Native Structure
To be Published
|
|
2C4U
 
 | | Crystal structure of the apo form of the 5'-Fluoro-5'-deoxyadenosine synthase enzyme from Streptomyces cattleya | | Descriptor: | 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, GLYCEROL | | Authors: | McEwan, A.R, Deng, H, Robinson, D.A, DeLaurentis, W, McGlinchey, R.P, O'Hagan, D, Naismith, J.H. | | Deposit date: | 2005-10-22 | | Release date: | 2006-04-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity in enzymatic fluorination. The fluorinase from Streptomyces cattleya accepts 2'-deoxyadenosine substrates.
Org. Biomol. Chem., 4, 2006
|
|
2C5B
 
 | | X-ray crystal structure of 5'-fluorodeoxyadenosine synthase from Streptomyces cattleya complexed with 2'deoxy-5'deoxy-fluoroadenosine. | | Descriptor: | 5'-FLUORO-2',5'-DIDEOXYADENOSINE, 5'-FLUORO-5'-DEOXYADENOSINE SYNTHASE, METHIONINE | | Authors: | McEwan, A.R, Deng, H, McGlinchey, R.P, Robinson, D.A, O'Hagan, D, Naismith, J.H, Spencer, J. | | Deposit date: | 2005-10-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate Specificity in Enzymatic Fluorination. The Fluorinase from Streptomyces Cattleya Accepts 2'-Deoxyadenosine Substrates.
Org.Biomol.Chem., 4, 2006
|
|
9I2R
 
 | | Wzc-K540M-3YE MgADP C8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative transmembrane protein Wzc | | Authors: | Liu, J.W, Yang, Y, Naismith, J.H. | | Deposit date: | 2025-01-22 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Molecular basis for the phosphorylation of bacterial tyrosine kinase Wzc.
Nat Commun, 16, 2025
|
|
2AQJ
 
 | | The structure of tryptophan 7-halogenase (PrnA) suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.-H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-18 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2AR8
 
 | | The structure of tryptophan 7-halogenase (PrnA)suggests a mechanism for regioselective chlorination | | Descriptor: | 7-CHLOROTRYPTOPHAN, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
