5UY1
 
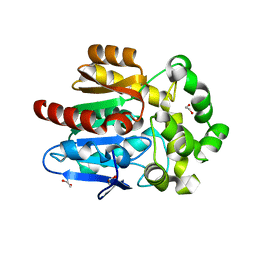 | | X-ray crystal structure of apo Halotag | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Cation-pi Interaction Enables a Halo-Tag Fluorogenic Probe for Fast No-Wash Live Cell Imaging and Gel-Free Protein Quantification.
Biochemistry, 56, 2017
|
|
5MQO
 
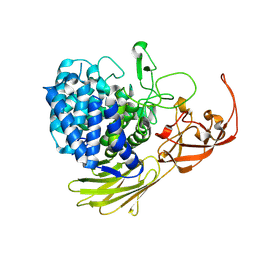 | | Glycoside hydrolase BT_1003 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
8ZMQ
 
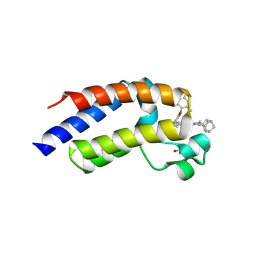 | | Crystal Structure of the second bromodomain of human BRD4 BD2 in complex with the inhibitor Y13190 | | Descriptor: | 2-(2-(adamantan-1-yl)-4-ethyl-1H-imidazol-5-yl)-7-(2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl)-5-methylfuro[3,2-c]pyridin-4(5H)-one, BRD4_HUMAN, DIMETHYL SULFOXIDE, ... | | Authors: | Li, J, Hu, Q, Xu, H, Zhao, X, Zhang, C, Zhu, R, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2024-05-23 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First BRD4 Second Bromodomain (BD2)-Selective Inhibitors.
J.Med.Chem., 67, 2024
|
|
8ZM8
 
 | | Crystal Structure of the first bromodomain of human BRD4 BD1 in complex with the inhibitor Y13221 | | Descriptor: | 2-[2-ethyl-4-(methoxymethyl)-1~{H}-imidazol-5-yl]-7-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-5-methyl-furo[3,2-c]pyridin-4-one, BRD4_HUMAN | | Authors: | Li, J, Hu, Q, Xu, H, Zhao, X, Zhang, C, Zhu, R, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2024-05-22 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of the First BRD4 Second Bromodomain (BD2)-Selective Inhibitors.
J.Med.Chem., 67, 2024
|
|
3C0F
 
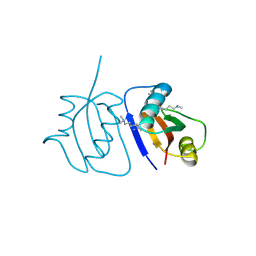 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
8ZMB
 
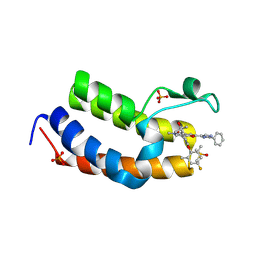 | | Crystal Structure of the first bromodomain of human BRD4 BD1 in complex with the inhibitor Y13195 | | Descriptor: | 7-(2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl)-5-methyl-2-(2-phenyl-1H-imidazol-5-yl)furo[3,2-c]pyridin-4(5H)-one, BRD4_HUMAN, DIMETHYL SULFOXIDE, ... | | Authors: | Li, J, Hu, Q, Xu, H, Zhao, X, Zhang, C, Zhu, R, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2024-05-22 | | Release date: | 2024-12-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of the First BRD4 Second Bromodomain (BD2)-Selective Inhibitors.
J.Med.Chem., 67, 2024
|
|
5ZYA
 
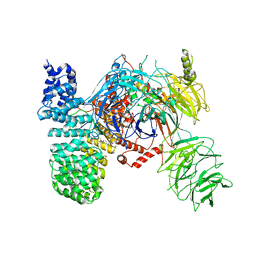 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
5LCV
 
 | |
9DI6
 
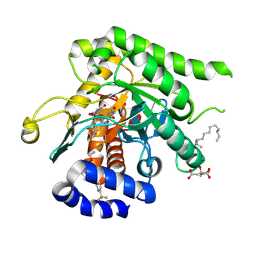 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM679 (ethyl 1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxylate) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, CITRIC ACID, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DIK
 
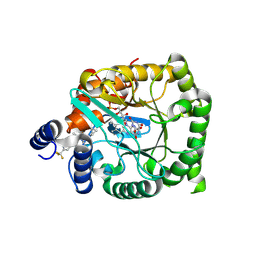 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM681 (N-cyclopropyl-1,4-dimethyl-5-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrazole-3-carboxamide) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-05 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DIZ
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM959 (6-cyclopropyl-2,4-dimethyl-3-((6-(trifluoromethyl)pyridin-3-yl)methyl)-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, 6-cyclopropyl-2,4-dimethyl-3-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-06 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1174 (3-((7-azaspiro[3.5]nonan-7-yl)methyl)-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 3-[(7-azaspiro[3.5]nonan-7-yl)methyl]-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-10 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DLY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1211 ((R)-3-(amino(6-(trifluoromethyl)pyridin-3-yl)methyl)-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 3-{(R)-amino[6-(trifluoromethyl)pyridin-3-yl]methyl}-4-cyclopropyl-6-ethyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchic, D, Phillips, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DLK
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1398 ((S)-3-(amino(3-chloro-4-(trifluoromethyl)phenyl)(cyclopropyl)methyl)-6-cyclopropyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one) | | Descriptor: | 3-[(R)-amino[3-chloro-4-(trifluoromethyl)phenyl](cyclopropyl)methyl]-6-cyclopropyl-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETIC ACID, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-11 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
9DKQ
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1153 (4,6-dicyclopropyl-3-(3-fluoro-4-(trifluoromethyl)benzyl)-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one) | | Descriptor: | 4,6-dicyclopropyl-3-{[3-fluoro-4-(trifluoromethyl)phenyl]methyl}-2-methyl-2,6-dihydro-7H-pyrazolo[3,4-d]pyridazin-7-one, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchic, D, Phillips, M. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-01 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
2ZO2
 
 | | Mouse NP95 SRA domain non-specific DNA complex | | Descriptor: | DNA (5'-D(*DAP*DAP*DCP*DTP*DGP*DCP*DGP*DCP*DAP*DGP*DTP*DT)-3'), E3 ubiquitin-protein ligase UHRF1, PHOSPHATE ION | | Authors: | Hashimoto, H, Horton, J.R, Cheng, X. | | Deposit date: | 2008-05-05 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix
Nature, 455, 2008
|
|
5LYY
 
 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 3-[2-(4-fluoranylphenoxy)ethyl]-1,3-diazaspiro[4.5]decane-2,4-dione, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
9DKO
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM1010 (6-cyclopropyl-2,4-dimethyl-3-(4-(trifluoromethyl)benzyl)-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one) | | Descriptor: | 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, 6-cyclopropyl-2,4-dimethyl-3-{[4-(trifluoromethyl)phenyl]methyl}-2,6-dihydro-7H-pyrazolo[3,4-c]pyridin-7-one, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Tomchick, D, Phillips, M. | | Deposit date: | 2024-09-09 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Based Discovery and Development of Highly Potent Dihydroorotate Dehydrogenase Inhibitors for Malaria Chemoprevention.
J.Med.Chem., 68, 2025
|
|
5MQS
 
 | | Sialidase BT_1020 | | Descriptor: | Beta-L-arabinobiosidase, CALCIUM ION, SODIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
8U8F
 
 | | GPR3 Orphan G-coupled Protein Receptor in complex with Dominant Negative Gs. | | Descriptor: | G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Russell, I.C, Belousoff, M.J, Sexton, P. | | Deposit date: | 2023-09-17 | | Release date: | 2024-03-06 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Lipid-Dependent Activation of the Orphan G Protein-Coupled Receptor, GPR3.
Biochemistry, 63, 2024
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6UVA
 
 | | CryoEM Structure of the active Adrenomedullin 2 receptor G protein complex with adrenomedullin 2 peptide | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
5MQR
 
 | | Sialidase BT_1020 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-L-arabinobiosidase, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
5MQP
 
 | | Glycoside hydrolase BT_1002 | | Descriptor: | CALCIUM ION, Glycoside hydrolase BT_1002 | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The most complex carbohydrate known is degraded in the human gut by single organisms and not bacterial consortia
To Be Published
|
|
5MT2
 
 | | Glycoside hydrolase BT_0996 | | Descriptor: | Beta-galactosidase, GLYCEROL | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
