4H3X
 
 | | Crystal structure of an MMP broad spectrum hydroxamate based inhibitor CC27 in complex with the MMP-9 catalytic domain | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
6HS5
 
 | | N-terminal domain including the conserved ImpA_N region of the TssA component of the type VI secretion system from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TssA | | Authors: | Dix, S.R, Owen, H.J, Sun, R, Ahmad, A, Shastri, S, Spiewak, H.L, Mosby, D.J, Harris, M.J, Batters, S.L, Brooker, T.A, Tzokov, S.B, Sedelnikova, S.E, Baker, P.J, Bullough, P.A, Rice, D.W, Thomas, M.S. | | Deposit date: | 2018-09-28 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the function of type VI secretion system TssA subunits.
Nat Commun, 9, 2018
|
|
4LJH
 
 | | Crystal Structure of Pseudomonas aeruginosa Lectin LecA Complexed with 1-Methyl-3-indolyl-b-D-galactopyranoside at 1.45 A Resolution | | Descriptor: | 1-methyl-1H-indol-3-ol, CALCIUM ION, PA-I galactophilic lectin, ... | | Authors: | Kadam, R.U, Stocker, A, Reymond, J.L. | | Deposit date: | 2013-07-04 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CH-pi "T-Shape" Interaction with Histidine Explains Binding of Aromatic Galactosides to Pseudomonas aeruginosa Lectin LecA
Acs Chem.Biol., 8, 2013
|
|
6HSD
 
 | | Crystal structure of the oxidized form of the transcription regulator RsrR | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-09-30 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Transcription Regulator RsrR Reveals a [2Fe-2S] Cluster Coordinated by Cys, Glu, and His Residues.
J. Am. Chem. Soc., 141, 2019
|
|
6H8N
 
 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis - mutant D285S | | Descriptor: | GLYCEROL, PHOSPHATE ION, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ... | | Authors: | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | Deposit date: | 2018-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
3ZTS
 
 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FINAL STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
6HTL
 
 | |
3ZCY
 
 | | Ascorbate peroxidase W41A-H42Y mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASCORBATE PEROXIDASE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2012-11-23 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Conformational Mobility of the Active Site of Ascorbate Peroxidase
Dalton Trans, 42, 2013
|
|
2CBE
 
 | |
6HAL
 
 | | Human carbonmonoxy hemoglobin SFX dataset | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Doak, B, Gorel, A, Foucar, L, Barends, T.R.M, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C.M, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrell, D.A, Owen, R.L, Schlichting, I. | | Deposit date: | 2018-08-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6H9Z
 
 | | Molecular bases of histo-blood group antigen recognition by the most common human rotavirus | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Outer capsid protein VP4, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Gozalbo-Rovira, R, Vila-Vicent, S, Buesa, J, Santiso-Bellon, C, Monedero, V, Yebra, M.J, Rodriguez-Diaz, J, Marina, A. | | Deposit date: | 2018-08-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unraveling the role of the secretor antigen in human rotavirus attachment to histo-blood group antigens.
Plos Pathog., 15, 2019
|
|
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
6HAS
 
 | |
6HBC
 
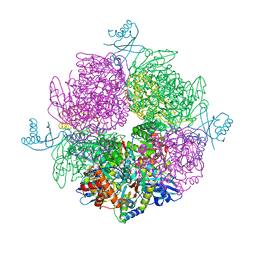 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HC2
 
 | | Crystal structure of NuMA/LGN hetero-hexamers | | Descriptor: | G-protein-signaling modulator 2, Nuclear mitotic apparatus protein 1 | | Authors: | Pasqualato, S, Culurgioni, S, Foadi, J, Alfieri, A, Mapelli, M. | | Deposit date: | 2018-08-13 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Hexameric NuMA:LGN structures promote multivalent interactions required for planar epithelial divisions.
Nat Commun, 10, 2019
|
|
5LYI
 
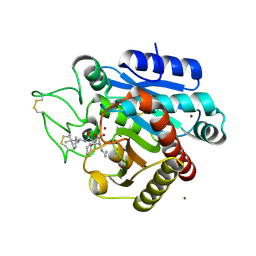 | | Crystal structure of 1 in complex with tafCPB | | Descriptor: | (2~{S})-6-azanyl-2-[[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(1~{R},2~{S},4~{R})-1,7,7-trimethyl-2-bicyclo[2.2.1]heptanyl]amino]propan-2-yl]sulfamoylamino]hexanoic acid, Carboxypeptidase B, ZINC ION | | Authors: | Schreuder, H, Liesum, A, Loenze, P. | | Deposit date: | 2016-09-28 | | Release date: | 2016-10-26 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Sulfamide as Zinc Binding Motif in Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (TAFIa).
J. Med. Chem., 59, 2016
|
|
5LZT
 
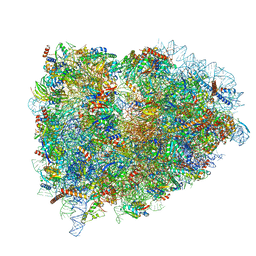 | | Structure of the mammalian ribosomal termination complex with eRF1 and eRF3. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Shao, S, Murray, J, Brown, A, Taunton, J, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2016-10-02 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes.
Cell, 167, 2016
|
|
3ZTR
 
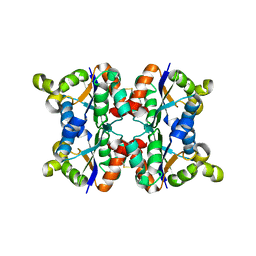 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FIRST STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
1RM5
 
 | | Crystal structure of mutant S188A of photosynthetic glyceraldehyde-3-phosphate dehydrogenase A4 isoform, complexed with NADP | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Sparla, F, Fermani, S, Falini, G, Ripamonti, A, Sabatino, P, Pupillo, P, Trost, P. | | Deposit date: | 2003-11-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Coenzyme Site-directed Mutants of Photosynthetic A(4)-GAPDH Show Selectively Reduced NADPH-dependent Catalysis, Similar to Regulatory AB-GAPDH Inhibited by Oxidized Thioredoxin
J.Mol.Biol., 340, 2004
|
|
2CBD
 
 | | STRUCTURE OF NATIVE AND APO CARBONIC ANHYDRASE II AND SOME OF ITS ANION-LIGAND COMPLEXES | | Descriptor: | CARBONIC ANHYDRASE II, SULFITE ION, ZINC ION | | Authors: | Hakansson, K, Carlsson, M, Svensson, L.A, Liljas, A. | | Deposit date: | 1992-06-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes.
J.Mol.Biol., 227, 1992
|
|
6JRE
 
 | | Structure of N-terminal domain of Plasmodium vivax p43 (PfNTD) solved by Co-SAD phasing | | Descriptor: | Aminoacyl-tRNA synthetase-interacting multifunctional protein p43, COBALT (II) ION | | Authors: | Manickam, Y, Harlos, K, Sharma, M, Gupta, S, Sharma, A. | | Deposit date: | 2019-04-03 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6JO3
 
 | | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with substrate sn-glycerol-1-phosphate | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Nemoto, N, Miyazono, K, Tanokura, M, Yamagishi, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with the substrate sn-glycerol 1-phosphate.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JOG
 
 | | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules. | | Descriptor: | ASCORBIC ACID, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Viswanathan, V, Gupta, A, Bairagya, H.R, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules.
To Be Published
|
|
1NQ6
 
 | | Crystal Structure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 | | Descriptor: | MAGNESIUM ION, Xys1 | | Authors: | Canals, A, Vega, M.C, Gomis-Ruth, F.X, Santamaria, R.I, Coll, M. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of xylanase Xys1delta from Streptomyces halstedii.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1RUM
 
 | | Crystal structure (F) of H2O2-soaked cationic cyclization antibody 4C6 fab at pH 8.5 with a data set collected at SSRL beamline 9-1. | | Descriptor: | BENZOIC ACID, GLYCEROL, immunoglobulin igg2a, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
