8GWE
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, MAGNESIUM ION, Non-structural protein 8, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWF
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.Y, Huang, Y.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWM
 
 | | SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP | | Descriptor: | 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate), Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWK
 
 | | SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
6IRO
 
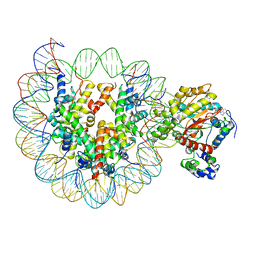 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2018-11-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6K1P
 
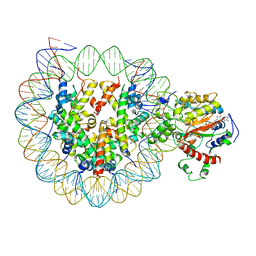 | | The complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JYL
 
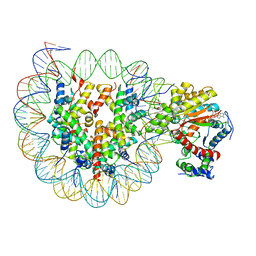 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2019-04-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7X8L
 
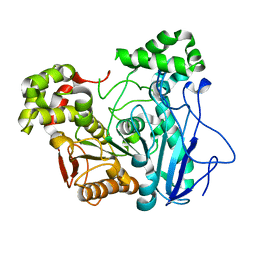 | |
8P5R
 
 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8Z0F
 
 | |
8Z9X
 
 | |
7ELM
 
 | | Structure of Csy-AcrIF24 | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7ELN
 
 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | 54-MER DNA, AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7DTR
 
 | | Structure of an AcrIF protein | | Descriptor: | AcrIF24 | | Authors: | Yue, F, Peipei, Y. | | Deposit date: | 2021-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
6V0A
 
 | | Crystal structure of cytochrome c nitrite reductase from the bacterium Geobacter lovleyi with bound sulfate | | Descriptor: | HEME C, Nitrite reductase (cytochrome; ammonia-forming), SULFATE ION | | Authors: | Satyanarayana, L, Campecino, J, Hegg, L.H, Hu, J. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cytochromecnitrite reductase from the bacteriumGeobacter lovleyirepresents a new NrfA subclass.
J.Biol.Chem., 295, 2020
|
|
8JIP
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIR
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIT
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIQ
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist Peptide 15-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIU
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIS
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist peptide15-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WE6
 
 | | Structure of Csy-AcrIF24-dsDNA | | Descriptor: | AcrIF24, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L, Feng, Y. | | Deposit date: | 2021-12-22 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into the inhibition of type I-F CRISPR-Cas system by a multifunctional anti-CRISPR protein AcrIF24.
Nat Commun, 13, 2022
|
|
7VB2
 
 | | Solution structure of human ribosomal protein uL11 | | Descriptor: | 60S ribosomal protein L12 | | Authors: | Lee, K.M, Wong, K.B. | | Deposit date: | 2021-08-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The flexible N-terminal motif of uL11 unique to eukaryotic ribosomes interacts with P-complex and facilitates protein translation.
Nucleic Acids Res., 50, 2022
|
|
6LHK
 
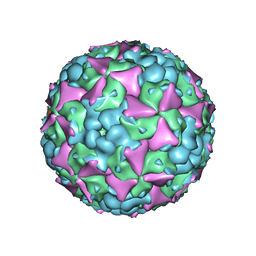 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHA
 
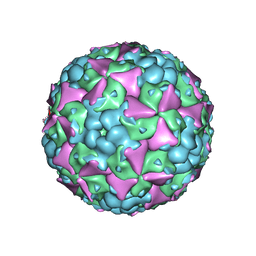 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
