3S1T
 
 | |
2VK1
 
 | | Crystal structure of the Saccharomyces cerevisiae pyruvate decarboxylase variant D28A in complex with its substrate | | Descriptor: | MAGNESIUM ION, PYRUVATE DECARBOXYLASE ISOZYME 1, PYRUVIC ACID, ... | | Authors: | Kutter, S, Weik, M, Weiss, M.S, Konig, S. | | Deposit date: | 2007-12-16 | | Release date: | 2009-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Covalently Bound Substrate at the Regulatory Site of Yeast Pyruvate Decarboxylases Triggers Allosteric Enzyme Activation.
J.Biol.Chem., 284, 2009
|
|
6Y08
 
 | | Mouse thymidylate synthase cocrystallized with dUMP and soaked in sulfamethoxazole | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Sulfamethoxazole, Thymidylate synthase | | Authors: | Maj, P, Jarmula, A, Wilk, P, Weiss, M.S, Rode, W. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Mouse thymidylate synthase cocrystallized with dUMP and soaked in sulfamethoxazole
To Be Published
|
|
8PH4
 
 | | Co-Crystal structure of the SARS-CoV2 main protease Nsp5 with an Uracil-carrying X77-like inhibitor | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, MALONATE ION, ... | | Authors: | Barthel, T, Altincekic, N, Jores, N, Wollenhaupt, J, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2023-06-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Targeting the Main Protease (M pro , nsp5) by Growth of Fragment Scaffolds Exploiting Structure-Based Methodologies.
Acs Chem.Biol., 19, 2024
|
|
8BOG
 
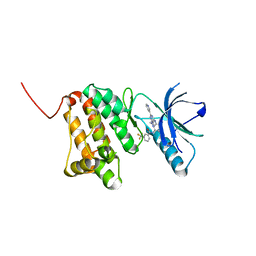 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 7 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-[4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOH
 
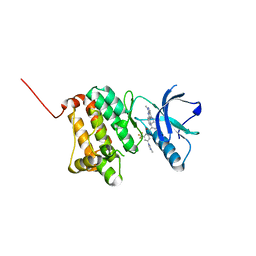 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 8 | | Descriptor: | 4-methyl-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide, Ephrin type-A receptor 2 | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOM
 
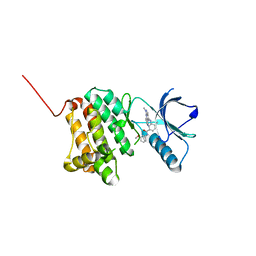 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 14 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3-methoxyphenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOD
 
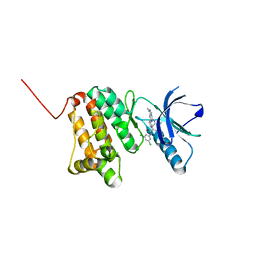 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 20 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-[3,5-bis(chloranyl)phenyl]-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOI
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 9 | | Descriptor: | 4-methyl-~{N}-[4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide, Ephrin type-A receptor 2 | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOK
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 11 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3,5-dimethoxyphenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOF
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 12 | | Descriptor: | 4-methyl-~{N}-(3-methylphenyl)-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide, Ephrin type-A receptor 2 | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
8BOC
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 19 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-[3,5-bis(trifluoromethyl)phenyl]-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of the Lead Compound NVP-BHG712 as a Colorectal Cancer Inhibitor.
Chemistry, 29, 2023
|
|
2JCW
 
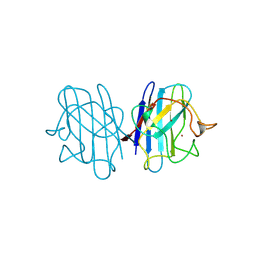 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (I) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-21 | | Release date: | 1999-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
7YD4
 
 | | Crystal structure of an N terminal truncated secreted protein, Rv0398c from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Possible secreted protein | | Authors: | Saha, R, Mukherjee, S, Singh, B.K, Weiss, M.S, De, S, Das, A.K. | | Deposit date: | 2022-07-03 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal structure of a mycobacterial secretory protein Rv0398c and in silico prediction of its export pathway.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
6TC2
 
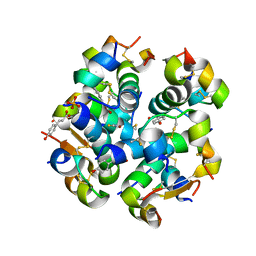 | | Monoclinic human insulin in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Insulin, PHOSPHATE ION, ... | | Authors: | Triandafillidis, D.-P, Parthenios, N, Spiliopoulou, M, Valmas, A, Kosinas, C, Gozzo, F, Reinle-Schmitt, M, Beckers, D, Degen, T, Pop, M, Fitch, A, Wollenhaupt, J, Weiss, M.S, Karavassili, F, Margiolaki, I. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Insulin polymorphism induced by two polyphenols: new crystal forms and advances in macromolecular powder diffraction.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6TTO
 
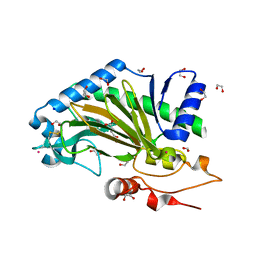 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FORMIC ACID, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6TTN
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, Hyoscyamine 6 beta-hydroxylase, N-OXALYLGLYCINE, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6ETE
 
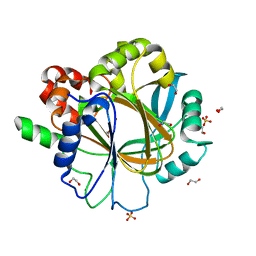 | | Crystal structure of KDM4D with tetrazolhydrazide compound 5 | | Descriptor: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, NICKEL (II) ION, ... | | Authors: | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | Deposit date: | 2017-10-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Crystal structure of KDM4D with tetrazolylhydrazide ligand NR128
To be published
|
|
6ETS
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6F5R
 
 | | Crystal Structure of KDM4D with GF028 ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-(3-oxidanylpropylamino)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.607 Å) | | Cite: | Crystal Structure of KDM4D with GF028 ligand
To be published
|
|
6F5S
 
 | | Crystal Structure of KDM4D with tetrazole ligand GF049 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure of KDM4D with tetrazole ligand GF049
To be published
|
|
6ETT
 
 | | Crystal structure of KDM4D with tetrazole compound 4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETV
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETU
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETW
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 3 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
