4PTS
 
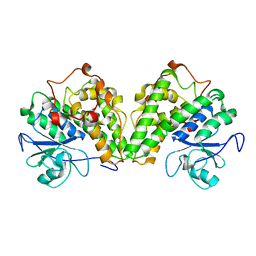 | | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405 | | Descriptor: | glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of a glutathione transferase from Gordonia bronchialis DSM 43247, target EFI-507405
To be Published
|
|
4PMJ
 
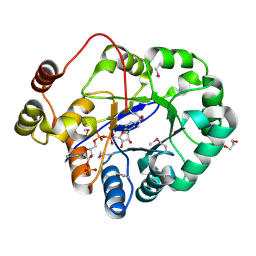 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Szlachta, K, Zimmerman, M.D, Hillerich, B.S, Gizzi, A, Toro, R, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-21 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Sinorhizobiummeliloti 1021 in complex with NADP
to be published
|
|
4PX1
 
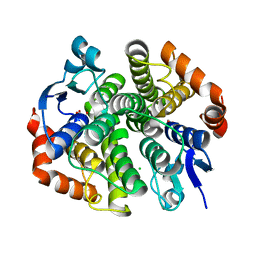 | | CRYSTAL STRUCTURE OF Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE (TARGET EFI-507068) | | Descriptor: | CHLORIDE ION, MALONIC ACID, Maleylacetoacetate isomerase (Glutathione S-transferase) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4MPF
 
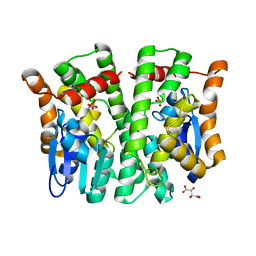 | | Crystal structure of human glutathione transferase theta-2, complex with inorganic phosphate, GSH free, target EFI-507257 | | Descriptor: | Glutathione S-transferase theta-2, L(+)-TARTARIC ACID, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Glutathione S-Transferase Theta-2 (Target Efi-507257)
To be Published
|
|
4NAX
 
 | | Crystal structure of glutathione transferase PPUT_1760 from Pseudomonas putida, target EFI-507288, with one glutathione disulfide bound per one protein subunit | | Descriptor: | FORMIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Crystal structure of glutathione transferase Pput_1760 from Pseudomonas putida, target EFI-507288
To be Published
|
|
4MZW
 
 | | CRYSTAL STRUCTURE OF NU-CLASS GLUTATHIONE TRANSFERASE YGHU FROM Streptococcus sanguinis SK36, COMPLEX WITH GLUTATHIONE DISULFIDE, TARGET EFI-507286 | | Descriptor: | ACETATE ION, Glutathione S-Transferase, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Yghu (Target Efi-507286)
To be Published
|
|
4PXO
 
 | | Crystal structure of Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE AND GSH (TARGET EFI-507068) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, MALONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from Methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4O7H
 
 | | Crystal structure of a glutathione S-transferase from Rhodospirillum rubrum F11, Target EFI-507460 | | Descriptor: | Glutathione S-transferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Stead, M, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Attonito, J.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-12-24 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a glutathione S-transferase from Rhodospirillum rubrum F11, Target EFI-507460
TO BE PUBLISHED
|
|
4Q2H
 
 | | Crystal structure of probable proline racemase from agrobacterium radiobacter K84, TARGET EFI-506561, with bound carbonate | | Descriptor: | BICARBONATE ION, GLYCEROL, Proline racemase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Proline Racemase Arad_0731 from Agrobacterium Radiobacter, Target Efi-506561
To be Published
|
|
4QS6
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 (TARGET EFI-505250) WITH BOUND 4-HYDROXY-3-METHOXY-5-NITROBENZOIC ACID, NO METAL, THE D314N MUTANT | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Ligw2 Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4QRN
 
 | | HIGH-RESOLUTION CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE (TARGET EFI-505250) FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 COMPLEXED WITH MANGANESE AND 4-HYDROXY-3-METHOXY-5-NITROBENZOIC ACID | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, 5-Carboxyvanillate Decarboxylase, ACETATE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Crystal Structure of 5-Carboxyvanillate Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4QS5
 
 | | CRYSTAL STRUCTURE of 5-CARBOXYVANILLATE DECARBOXYLASE LIGW2 FROM NOVOSPHINGOBIUM AROMATICIVORANS DSM 12444 (TARGET EFI-505250) WITH BOUND MANGANESE AND 3-methoxy-4-hydroxy-5-nitrobenzoic acid, THE D314N MUTANT | | Descriptor: | 4-hydroxy-3-methoxy-5-nitrobenzoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Raushel, M, Almo, S.C. | | Deposit date: | 2014-07-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 5-CARBOXYVANILLATE Decarboxylase from Novosphingobium Aromaticivorans
To be Published
|
|
4QRO
 
 | | CRYSTAL STRUCTURE of DIHYDROXYBENZOIC ACID DECARBBOXYLASE BPRO_2061 (TARGET EFI-500288) FROM POLAROMONAS SP. JS666 WITH BOUND MANGANESE AND AN INHIBITOR, 2-NITRORESORCINOL | | Descriptor: | 2-nitrobenzene-1,3-diol, ACETATE ION, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Vladimirova, A, Toro, R, Bhosle, R, Gerlt, J.A, Raushel, M, Almo, S.C. | | Deposit date: | 2014-07-01 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Dihydroxybenzoate Decarboxylase from Frompolaromonas Sp WITH BOUND MANGANESE AND 2-NITRORESORCINOL
To be Published
|
|
4R2F
 
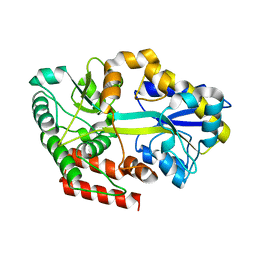 | | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus A6, target EFI-510633, with bound laminaribiose | | Descriptor: | Extracellular solute-binding protein family 1, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-11 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus, target EFI-510633
To be Published
|
|
4QSE
 
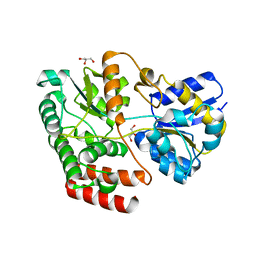 | | Crystal structure of ATU4361 sugar transporter from Agrobacterium Fabrum c58, target efi-510558, with bound glycerol | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, GLYCEROL | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, K.L, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of maltoside transporter ATU4361 from
Agrobacterium Fabrum, target EFI-510558
To be Published
|
|
4R6Y
 
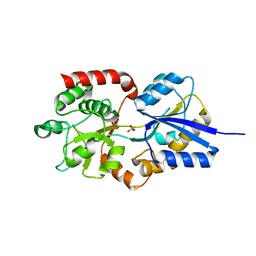 | | Crystal structure of solute-binding protein stm0429 from salmonella enterica subsp. enterica serovar typhimurium str. lt2, target efi-510776, a closed conformation, in complex with glycerol and acetate | | Descriptor: | ACETATE ION, GLYCEROL, Putative 2-aminoethylphosphonate-binding periplasmic protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of Transporter STM0429 from Salmonella Enterica
To be Published
|
|
4QSC
 
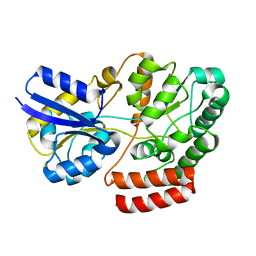 | | Crystal structure of ATU4361 sugar transporter from Agrobacterium Fabrum C58, target efi-510558, with bound maltose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Maltoside Transporter Atu4361 from Agrobacterium Fabrum, Target Efi-510558
To be Published
|
|
3OA4
 
 | | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125 | | Descriptor: | GLYCEROL, SULFATE ION, ZINC ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Garrett, S, Foti, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125
To be Published
|
|
3SN4
 
 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bonanno, J.B, Patskovsky, Y, Toro, R, Dickey, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium and alpha-ketoglutarate
To be Published
|
|
2O8R
 
 | | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis | | Descriptor: | Polyphosphate kinase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-12 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Polyphosphate Kinase from Porphyromonas Gingivalis
To be Published
|
|
2O3J
 
 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
2NX2
 
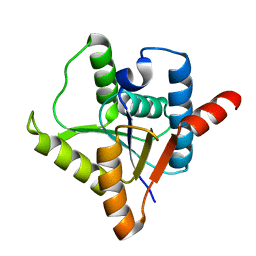 | | Crystal structure of protein ypsA from Bacillus subtilis, Pfam DUF1273 | | Descriptor: | Hypothetical protein ypsA | | Authors: | Ramagopal, U.A, Alvarado, J, Dickey, M, Reyes, C, Toro, R, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-16 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of hypothetical protein ypsA from Bacillus subtilis.
To be Published
|
|
2O34
 
 | | Crystal structure of protein DVU1097 from Desulfovibrio vulgaris Hildenborough, Pfam DUF375 | | Descriptor: | Hypothetical protein, SODIUM ION | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Hypothetical Protein from Desulfovibrio vulgaris Hildenborough
To be Published
|
|
2OGF
 
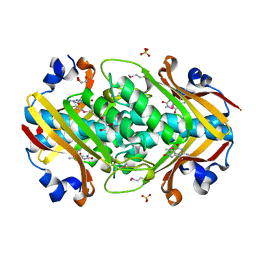 | | Crystal structure of protein MJ0408 from Methanococcus jannaschii, Pfam DUF372 | | Descriptor: | 8-OXOGUANINE, GLYCEROL, Hypothetical protein MJ0408, ... | | Authors: | Ramagopal, U.A, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-05 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of hypothetical protein MJ0408 from Methanococcus jannaschii
To be Published
|
|
2NQL
 
 | | Crystal structure of a member of the enolase superfamily from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, Isomerase/lactonizing enzyme, SODIUM ION, ... | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-31 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Enolase from Agrobacterium Tumefaciens C58
To be Published
|
|
