5R46
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, room temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R43
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 7.5, cryo temperature | | Descriptor: | Chymotrypsinogen A, IODIDE ION, MALONIC ACID, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4B
 
 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R4D
 
 | | Crystal Structure of gamma-Chymotrypsin at pH 9, cryo temperature | | Descriptor: | IODIDE ION, SULFATE ION, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5R47
 
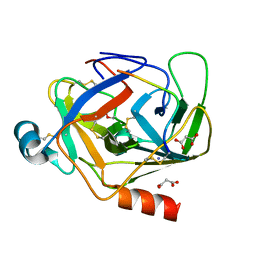 | | Crystal Structure of deuterated gamma-Chymotrypsin at pH 5.6, cryo temperature | | Descriptor: | IODIDE ION, MALONIC ACID, gamma-chymotrypsin, ... | | Authors: | Kreinbring, C.A, Wilson, M.A, Kovalevsky, A.Y, Blakeley, M.P, Fisher, S.Z, Lazar, L.M, Moulin, A.G, Novak, W.R, Petsko, G.A, Ringe, D. | | Deposit date: | 2020-02-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Effect of Temperature and pH on Ionizable Residues in gamma-Chymotrypsin: a X-ray and Neutron Crystallography Study
To be published
|
|
5RHN
 
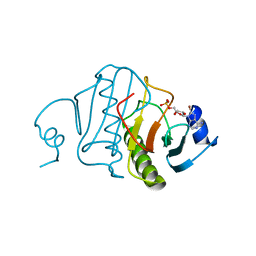 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT COMPLEXED WITH 8-BR-AMP | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-26 | | Release date: | 1997-06-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
5RAT
 
 | |
5T4J
 
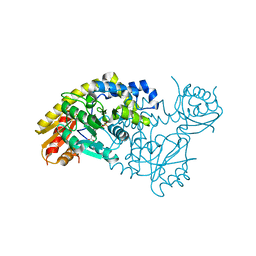 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, HTH-type transcriptional regulatory protein GabR, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4K
 
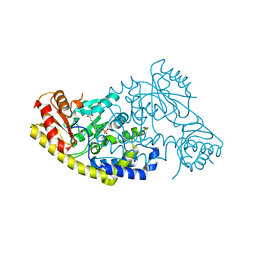 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4S)-5-fluoro-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, HTH-type transcriptional regulatory protein GabR | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4L
 
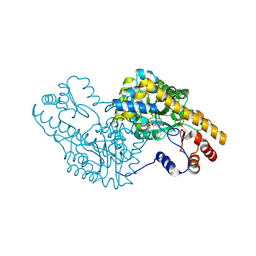 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T1J
 
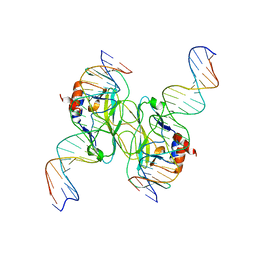 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | Descriptor: | DNA, T-box transcription factor TBX21 | | Authors: | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5UCD
 
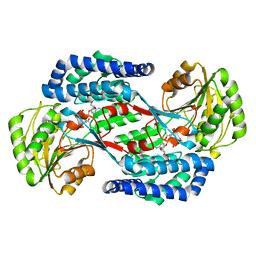 | | Benzaldehyde Dehydrogenase, a Class 3 Aldehyde Dehydrogenase, with bound NADP+ and Benzoate Adduct | | Descriptor: | NAD(P)-dependent benzaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zahniser, M.P.D, Prasad, S, Kneen, M.M, Kreinbring, C.A, Petsko, G.A, Ringe, D, McLeish, M.J. | | Deposit date: | 2016-12-22 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure and mechanism of benzaldehyde dehydrogenase from Pseudomonas putida ATCC 12633, a member of the Class 3 aldehyde dehydrogenase superfamily.
Protein Eng. Des. Sel., 30, 2017
|
|
1CNZ
 
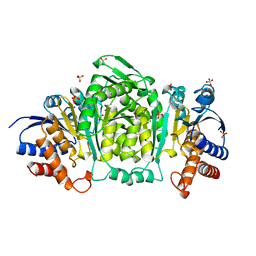 | | 3-ISOPROPYLMALATE DEHYDROGENASE (IPMDH) FROM SALMONELLA TYPHIMURIUM | | Descriptor: | MANGANESE (II) ION, PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE), SULFATE ION | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-24 | | Release date: | 1999-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of Eschericia Coli and Salmonella Typhimurium 3-
Isopropylmalate Dehydrogenase and Comparison with Their Thermophilic
Counterpart from Thermus Thermophilus
J.Mol.Biol., 266, 1997
|
|
1ESA
 
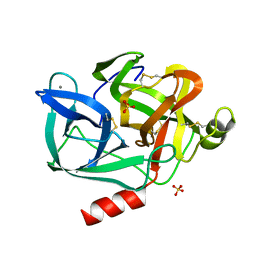 | | DIRECT STRUCTURE OBSERVATION OF AN ACYL-ENZYME INTERMEDIATE IN THE HYDROLYSIS OF AN ESTER SUBSTRATE BY ELASTASE | | Descriptor: | CALCIUM ION, PORCINE PANCREATIC ELASTASE, SULFATE ION | | Authors: | Ding, X, Rasmussen, B, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-04 | | Release date: | 1994-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct structural observation of an acyl-enzyme intermediate in the hydrolysis of an ester substrate by elastase.
Biochemistry, 33, 1994
|
|
1DQR
 
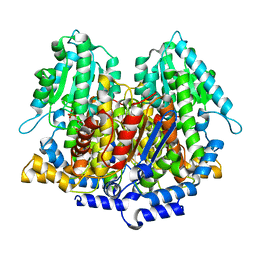 | | CRYSTAL STRUCTURE OF RABBIT PHOSPHOGLUCOSE ISOMERASE, A GLYCOLYTIC ENZYME THAT MOONLIGHTS AS NEUROLEUKIN, AUTOCRINE MOTILITY FACTOR, AND DIFFERENTIATION MEDIATOR | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Bahnson, B.J, Jeffery, C.J, Ringe, D, Petsko, G.A. | | Deposit date: | 2000-01-05 | | Release date: | 2000-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rabbit phosphoglucose isomerase, a glycolytic enzyme that moonlights as neuroleukin, autocrine motility factor, and differentiation mediator.
Biochemistry, 39, 2000
|
|
1DZ9
 
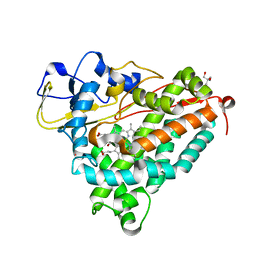 | | Putative oxo complex of P450cam from Pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ4
 
 | | ferric p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-16 | | Release date: | 2000-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ6
 
 | | ferrous p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ8
 
 | | oxygen complex of p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1CP6
 
 | | 1-BUTANEBORONIC ACID BINDING TO AEROMONAS PROTEOLYTICA AMINOPEPTIDASE | | Descriptor: | 1-BUTANE BORONIC ACID, PROTEIN (AMINOPEPTIDASE), ZINC ION | | Authors: | Depaola, C.C, Bennett, B, Holz, R.C, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-06-08 | | Release date: | 1999-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-Butaneboronic acid binding to Aeromonas proteolytica aminopeptidase: a case of arrested development.
Biochemistry, 38, 1999
|
|
1ELB
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-leucinamide, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1CM7
 
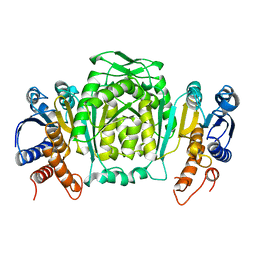 | | 3-ISOPROPYLMALATE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (3-ISOPROPYLMALATE DEHYDROGENASE) | | Authors: | Wallon, G, Kryger, G, Lovett, S.T, Oshima, T, Ringe, D, Petsko, G.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of Escherichia coli and Salmonella typhimurium 3-isopropylmalate dehydrogenase and comparison with their thermophilic counterpart from Thermus thermophilus.
J.Mol.Biol., 266, 1997
|
|
1ELC
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-phenylalaninamide, CALCIUM ION, ELASTASE | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1EY3
 
 | |
1BKH
 
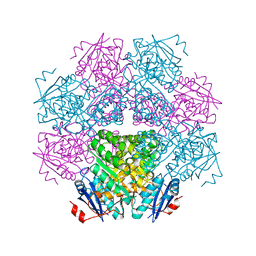 | | MUCONATE LACTONIZING ENZYME FROM PSEUDOMONAS PUTIDA | | Descriptor: | MUCONATE LACTONIZING ENZYME | | Authors: | Hasson, M.S, Schlichting, I, Moulai, J, Taylor, K, Barrett, W, Kenyon, G.L, Babbitt, P.C, Gerlt, J.A, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-07-07 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
