7RI9
 
 | | The structure of BAM in MSP1E3D1 at 6.9 Angstrom resolution | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI5
 
 | | Structure of a BAM in MSP1E3D1 nanodiscs at 4 Angstrom resolution | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RJ5
 
 | | The structure of BAM in complex with EspP at 7 Angstrom resolution | | Descriptor: | Maltodextrin-binding protein,Autotransporter outer membrane beta-barrel domain-containing protein chimera, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-20 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI8
 
 | | The structure of BAM in MSP2N2 nanodiscs | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI6
 
 | |
7RI4
 
 | | Structure of a BAM/EspP(beta9-12) hybrid-barrel intermediate | | Descriptor: | EspPbeta9-12, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7RI7
 
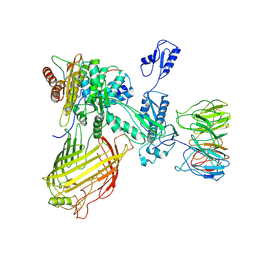 | | The structure of BAM in MSP1D1 nanodiscs | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Wu, R.R, Noinaj, N. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Plasticity within the barrel domain of BamA mediates a hybrid-barrel mechanism by BAM.
Nat Commun, 12, 2021
|
|
7YGI
 
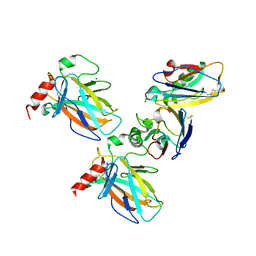 | | Crystal structure of p53 DBD domain in complex with azurin | | Descriptor: | Azurin, Cellular tumor antigen p53, PHOSPHATE ION, ... | | Authors: | Jiang, W.X, Zuo, J.Q, Hu, J.J, Chen, X.Q, Ma, L.X, Liu, Z, Xing, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of bacterial effector protein azurin targeting tumor suppressor p53 and inhibiting its ubiquitination.
Commun Biol, 6, 2023
|
|
6LS5
 
 | | Structure of human liver FBPase complexed with covalent allosteric inhibitor | | Descriptor: | 2-(ethyldisulfanyl)-1,3-benzothiazole, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Rongrong, S, Yixiang, X, Shuaishuai, N, Yanliang, R, Jian, L, Jian, W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Identification of the New Covalent Allosteric Binding Site of Fructose-1,6-bisphosphatase with Disulfiram Derivatives toward Glucose Reduction.
J.Med.Chem., 63, 2020
|
|
3MEF
 
 | |
5WK1
 
 | |
5ZBZ
 
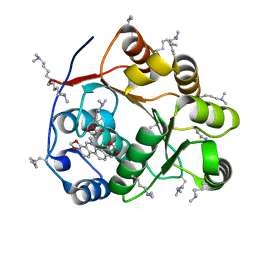 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
6CQ1
 
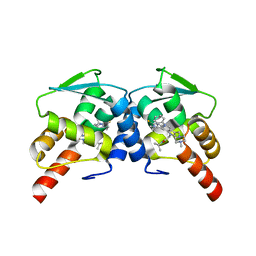 | | BCL6 BTB domain in complex with 15a | | Descriptor: | 2-{6-({[2-(1H-indol-3-yl)ethyl]carbamothioyl}amino)-3-[(4-methylpiperazin-1-yl)methyl]-1H-indol-1-yl}-N-(propan-2-yl)acetamide, B-cell lymphoma 6 protein | | Authors: | Linhares, B.M, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69921041 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
7BP6
 
 | |
7BP2
 
 | |
7BP5
 
 | |
7BP4
 
 | |
6C3L
 
 | | Crystal structure of BCL6 BTB domain with compound 15f | | Descriptor: | B-cell lymphoma 6 protein, N-[2-(1H-indol-3-yl)ethyl]-N'-{3-[(4-methylpiperazin-1-yl)methyl]-1-[2-(morpholin-4-yl)-2-oxoethyl]-1H-indol-6-yl}thiourea | | Authors: | Linhares, B, Cheng, H, Cierpicki, T, Xue, F. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.46092153 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6C3N
 
 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
4O1I
 
 | | Crystal Structure of the regulatory domain of MtbGlnR | | Descriptor: | Transcriptional regulatory protein | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
4O1H
 
 | | Crystal Structure of the regulatory domain of AmeGlnR | | Descriptor: | Transcription regulator GlnR | | Authors: | Lin, W, Wang, C, Zhang, P. | | Deposit date: | 2013-12-16 | | Release date: | 2014-04-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atypical OmpR/PhoB Subfamily Response Regulator GlnR of Actinomycetes Functions as a Homodimer, Stabilized by the Unphosphorylated Conserved Asp-focused Charge Interactions
J.Biol.Chem., 289, 2014
|
|
