2JC7
 
 | |
8R6M
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to I-BET151 | | Descriptor: | 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, Candida glabrata strain CBS138 chromosome C complete sequence, GLYCEROL, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6I
 
 | |
8R6K
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a phenyltriazine ligand | | Descriptor: | 6-methyl-~{N}-[(5-methylfuran-2-yl)methyl]-3-(4-methylphenyl)-1,2,4-triazin-5-amine, Candida glabrata strain CBS138 chromosome C complete sequence, PHOSPHATE ION, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6N
 
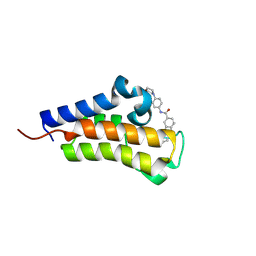 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to a pyridoindole ligand | | Descriptor: | 2-ethanoyl-~{N}-(4-morpholin-4-ylphenyl)-1,3,4,5-tetrahydropyrido[4,3-b]indole-8-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6L
 
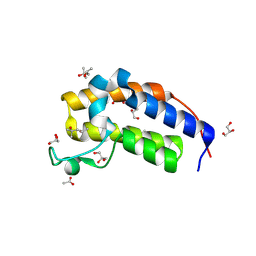 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 in the unbound state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Candida glabrata strain CBS138 chromosome C complete sequence, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6J
 
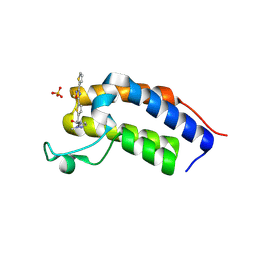 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a pyrazole ligand | | Descriptor: | 2-methyl-~{N}-[[5-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)thiophen-2-yl]methyl]pyrazole-3-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence, SULFATE ION | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
7BHD
 
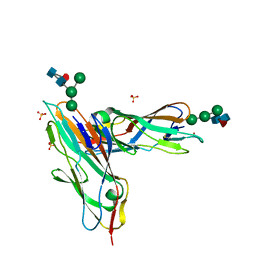 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2021-01-11 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
7QUO
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
7NYL
 
 | | Mutant H493A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | SH3 domain of JNK-interacting Protein 1 (JIP1), TETRAETHYLENE GLYCOL, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZD
 
 | | Fourth SH3 domain of POSH (Plenty of SH3 Domains protein) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase SH3RF1 | | Authors: | Palencia, A, Bessa, L.M, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYO
 
 | | Mutant A541L of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, SH3 domain of JNK-interacting Protein 1 (JIP1), SULFATE ION, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZC
 
 | |
7NYK
 
 | |
7NYM
 
 | | Mutant V517A - SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | HEXAETHYLENE GLYCOL, PHOSPHATE ION, SH3 domain of JNK-interacting Protein 1 (JIP1), ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NZB
 
 | | Mutant V517L of the SH3 domain of JNK-interacting protein 1 (JIP1) | | Descriptor: | PHOSPHATE ION, SH3 domain of JNK-interacting protein 1 (JIP1), TETRAETHYLENE GLYCOL | | Authors: | Perez, L.M, Ielasi, F.S, Jensen, M.R, Palencia, A. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
7NYN
 
 | | Mutant Y526A of SH3 domain of JNK-interacting Protein 1 (JIP1) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Perez, L.M, Ielasi, F.S, Palencia, A, Jensen, M.R. | | Deposit date: | 2021-03-23 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Visualizing protein breathing motions associated with aromatic ring flipping.
Nature, 602, 2022
|
|
8BY3
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-12-11 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.186 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
8R7H
 
 | | Cryo-EM structure of Human SHMT1 | | Descriptor: | Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2023-11-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 84, 2024
|
|
8BXY
 
 | | FimH in complex with alpha1,6 core-fucosylated oligomannose-3, crystallized in the trigonal space group | | Descriptor: | NICKEL (II) ION, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin, ... | | Authors: | Bridot, C, Bouckaert, J, Krammer, E.-M. | | Deposit date: | 2022-12-11 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors.
J.Biol.Chem., 299, 2023
|
|
1A7N
 
 | |
1A7P
 
 | |
1A7Q
 
 | | FV FRAGMENT OF MOUSE MONOCLONAL ANTIBODY D1.3 (BALB/C, IGG1, K) HIGH AFFINITY EXPRESSED VARIANT CONTAINING SER26L->GLY, ILE29L->THR, GLU81L->ASP, THR97L->SER, PRO240H->LEU, ASP258H->ALA, LYS281H->GLU, ASN283H->ASP AND LEU312H->VAL | | Descriptor: | IGG1-KAPPA D1.3 FV (HEAVY CHAIN), IGG1-KAPPA D1.3 FV (LIGHT CHAIN) | | Authors: | Marks, C, Henrick, K, Winter, G. | | Deposit date: | 1998-03-16 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structures of D1.3 Fv Mutants
To be Published
|
|
1A7O
 
 | |
6SBO
 
 | | Estrogen receptor mutant L536S | | Descriptor: | 6-(2,4-dichlorophenyl)-5-[4-[(3~{S})-1-(3-fluoranylpropyl)pyrrolidin-3-yl]oxyphenyl]-8,9-dihydro-7~{H}-benzo[7]annulene-2-carboxylic acid, Estrogen receptor | | Authors: | Vallee, F, Steier, V, Rak, A. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Discovery of 6-(2,4-Dichlorophenyl)-5-[4-[(3S)-1-(3-fluoropropyl)pyrrolidin-3-yl]oxyphenyl]-8,9-dihydro-7H-benzo[7]annulene-2-carboxylic acid (SAR439859), a Potent and Selective Estrogen Receptor Degrader (SERD) for the Treatment of Estrogen-Receptor-Positive Breast Cancer.
J.Med.Chem., 63, 2020
|
|
