3QKR
 
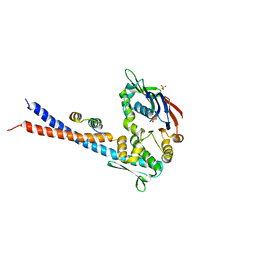 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, PHOSPHATE ION | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1ZGO
 
 | |
1XDC
 
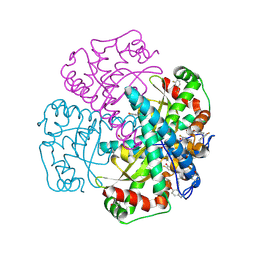 | | Hydrogen Bonding in Human Manganese Superoxide Dismutase containing 3-Fluorotyrosine | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Ayala, I, Perry, J.J, Szczepanski, J, Cabelli, D.E, Tainer, J.A, Vala, M.T, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Hydrogen bonding in human manganese superoxide dismutase containing 3-fluorotyrosine
Biophys.J., 89, 2005
|
|
1XIL
 
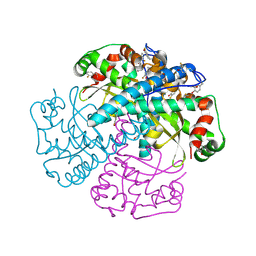 | | HYDROGEN BONDING IN HUMAN MANGANESE SUPEROXIDE DISMUTASE CONTAINING 3-FLUOROTYROSINE | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Ayala, I, Perry, J.J, Szczepanski, J, Cabelli, D.E, Tainer, J.A, Vala, M.T, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Hydrogen bonding in human manganese superoxide dismutase containing 3-fluorotyrosine
Biophys.J., 89, 2005
|
|
1ZGQ
 
 | |
3QKT
 
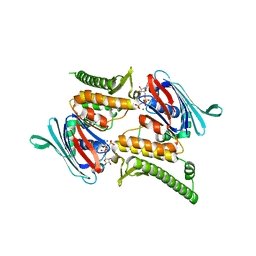 | | Rad50 ABC-ATPase with adjacent coiled-coil region in complex with AMP-PNP | | Descriptor: | DNA double-strand break repair rad50 ATPase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2R27
 
 | | Constitutively zinc-deficient mutant of human superoxide dismutase (SOD), C6A, H80S, H83S, C111S | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn] | | Authors: | Roberts, B.R, Getzoff, E.D, Karplus, P.A, Beckman, J.S, Tainer, J.A. | | Deposit date: | 2007-08-24 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of zinc-deficient human superoxide dismutase and implications for ALS.
J.Mol.Biol., 373, 2007
|
|
2QT2
 
 | | Cyclized-Dehydrated Intermediate of GFP Variant Q183E in Chromophore Maturation | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-07-31 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetically Isolated Reaction Intermediates Provide Structural Characterization of the Green Fluorescence Protein Fluorophore Biosynthesis Pathway
To be Published
|
|
2QRF
 
 | | Green Fluorescent Protein: Cyclized-only Intermediate of Chromophore Maturation in the Q183E variant | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, MAGNESIUM ION | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-07-28 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Green Fluorescent Protein Chromophore - Cyclized Intermediate
To be Published
|
|
2QZ0
 
 | | Mature Q183E variant of Green Fluorescent Protein Chromophore | | Descriptor: | Green fluorescent protein, MAGNESIUM ION | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-08-15 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Kinetically Isolated Reaction Intermediates Provide Structural Characterization of the GFP Fluorophore Biosynthesis Pathway
To be Published
|
|
2UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH H187D MUTANT UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2OD8
 
 | | Structure of a peptide derived from Cdc9 bound to PCNA | | Descriptor: | DNA ligase I, mitochondrial precursor, Proliferating cell nuclear antigen | | Authors: | Chapados, B.R, Tainer, J.A. | | Deposit date: | 2006-12-21 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The C-terminal domain of yeast PCNA is required for physical and functional interactions with Cdc9 DNA ligase.
Nucleic Acids Res., 35, 2007
|
|
2UGI
 
 | | PROTEIN MIMICRY OF DNA FROM CRYSTAL STRUCTURES OF THE URACIL GLYCOSYLASE INHIBITOR PROTEIN AND ITS COMPLEX WITH ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | Descriptor: | IMIDAZOLE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-11-06 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
3CRV
 
 | | XPD_Helicase | | Descriptor: | CITRATE ANION, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3CRW
 
 | | XPD_APO | | Descriptor: | HEXACYANOFERRATE(3-), XPD/Rad3 related DNA helicase | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations.
Cell(Cambridge,Mass.), 133, 2008
|
|
3DSC
 
 | | Crystal structure of P. furiosus Mre11 DNA synaptic complex | | Descriptor: | DNA (5'-D(P*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DAP*DC)-3'), DNA double-strand break repair protein mre11 | | Authors: | Williams, R.S, Moncalian, G, Shin, D.S, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3E6L
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C132283 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ETHYL 4-[(4-CHLOROPYRIDIN-2-YL)AMINO]PIPERIDINE-1-CARBOXYLATE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E7I
 
 | | Structure of murine inos oxygenase domain with inhibitor AR-C94864 | | Descriptor: | (2R)-5-FLUORO-2-(2-THIENYL)-1,2-DIHYDROQUINAZOLIN-4-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E7T
 
 | | Structure of murine iNOS oxygenase domain with inhibitor AR-C102222 | | Descriptor: | 1-(6-CYANO-3-PYRIDYLCARBONYL)-5',8'-DIFLUOROSPIRO[PIPERIDINE-4,2'(1'H)-QUINAZOLINE]-4'-AMINE, 7,8-DIHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-18 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E67
 
 | | Murine inos dimer with inhibitor 4-MAP bound | | Descriptor: | 4-METHYLPYRIDIN-2-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3E6T
 
 | | Structure of murine INOS oxygenase domain with inhibitor AR-C118901 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 5-(4'-AMINO-1'-ETHYL-5',8'-DIFLUORO-1'H-SPIRO[PIPERIDINE-4,2'-QUINAZOLINE]-1-YLCARBONYL)PICOLINONITRILE, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stueh, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3DSD
 
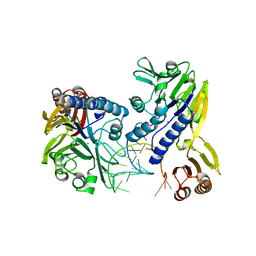 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | Authors: | Williams, R.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
3EBF
 
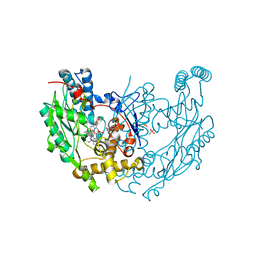 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | (3R)-3-[(1,2,3,4-tetrahydroisoquinolin-7-yloxy)methyl]-2,3-dihydrothieno[2,3-f][1,4]oxazepin-5-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3EBD
 
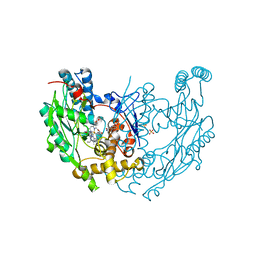 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | (2S)-2-methyl-2,3-dihydrothieno[2,3-f][1,4]oxazepin-5-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-27 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3ENM
 
 | | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 6, GLYCEROL, ... | | Authors: | Min, X, Akella, R, He, H, Humphreys, J.M, Tsutakawa, S, Lee, S.-J, Tainer, J.A, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-03-03 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the MAP2K MEK6 reveals an autoinhibitory dimer
Structure, 17, 2009
|
|
