5JJV
 
 | |
7NRG
 
 | |
7NWR
 
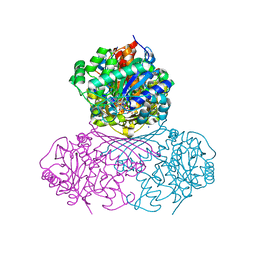 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
3R9H
 
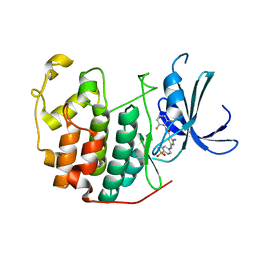 | | CDK2 in complex with inhibitor RC-2-142 | | Descriptor: | 4-amino-N-(2,6-difluorophenyl)-2-[(4-sulfamoylphenyl)amino]-1,3-thiazole-5-carboxamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-25 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
7NWY
 
 | |
2I9E
 
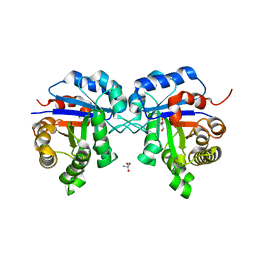 | | Structure of Triosephosphate Isomerase of Tenebrio molitor | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Triosephosphate isomerase | | Authors: | Schmidt, A, Scheerer, P, Wessner, H, Hoehne, W, Krauss, N. | | Deposit date: | 2006-09-05 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A coleopteran triosephosphate isomerase: X-ray structure and phylogenetic impact of insect sequences.
Insect Mol Biol, 19, 2010
|
|
7O2M
 
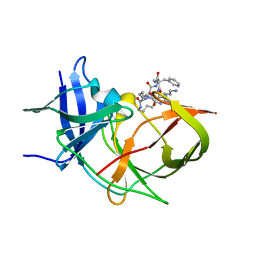 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2289 | | Descriptor: | 1-[(3~{S},6~{S},9~{S},19~{R})-3,6-bis(4-azanylbutyl)-2,5,8,12,15,18-hexakis(oxidanylidene)-9-(phenylmethyl)-1,4,7,11,14,17-hexazacyclotricos-19-yl]guanidine, Genome polyprotein | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7OC2
 
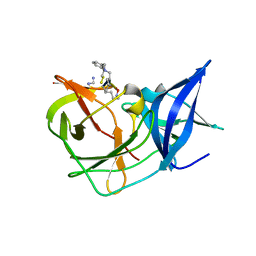 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2295 | | Descriptor: | Cyclic 1[2-CHLORO-4-METHOXY-PHENYL-OXYMETHYL]-4-[2,6-DICHLORO-PHENYL-OXYMETHYL]-BENZENE-(7-3)-7-BENZYL-1,3-DIMETHYL-8-PIPERAZIN-1-YL-3,7-DIHYDRO-PURINE-2,6-DIONE-(7-19)-N-ACETYL-L-CYSTEINE-(8-25)-[3R-[3A,4A,5B(S*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID-()-(6E,11E)-HEPTADECA-6,11-DIENE-9,9-DIYLBIS(PHOSPHONIC ACID), Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Heine, A, Steinmetzer, T. | | Deposit date: | 2021-04-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
7OD2
 
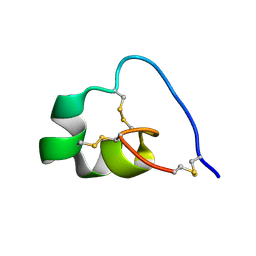 | |
7O55
 
 | |
4QQZ
 
 | | Crystal structure of T. fusca Cas3-AMPPNP | | Descriptor: | CRISPR-associated helicase, Cas3 family, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5JHJ
 
 | | M. Oryzae effector AVR-Pia mutant H3 | | Descriptor: | Antivirulence protein AVR-Pia | | Authors: | Padilla, A, deGuillen, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the Magnaporthe oryzae Effector AVR-Pia by the Decoy Domain of the Rice NLR Immune Receptor RGA5.
Plant Cell, 29, 2017
|
|
7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
7OBV
 
 | |
7O3N
 
 | | Crystal Structure of AcrB Single Mutant - 2 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Ababou, A. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.561 Å) | | Cite: | Crystal Structure of AcrB Single Mutant - 2
To Be Published
|
|
7O04
 
 | |
4QVS
 
 | | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405 | | Descriptor: | CHLORIDE ION, S-layer domain-containing protein, SODIUM ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of S-layer domain-containing protein (residues 221-444) from Clostridium thermocellum ATCC 27405
To be Published
|
|
4QQY
 
 | | Crystal structure of T. fusca Cas3-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CRISPR-associated helicase, Cas3 family, ... | | Authors: | Ke, A, Huo, Y, Nam, K.H. | | Deposit date: | 2014-06-30 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7NZT
 
 | | Crystal structure of chimeric carbonic anhydrase VA with 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-(cyclooctylamino)-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-03-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of chimeric carbonic anhydrase VA
To Be Published
|
|
4QX4
 
 | | Human Aldose Reductase complexed with a ligand with a new scaffold at 1.26 A | | Descriptor: | (3-thioxo-2,3-dihydro-5H-[1,2,4]triazino[5,6-b]indol-5-yl)acetic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Rechlin, C, Heine, A, Klebe, G. | | Deposit date: | 2014-07-18 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.259 Å) | | Cite: | Identification of novel aldose reductase inhibitors based on carboxymethylated mercaptotriazinoindole scaffold.
J.Med.Chem., 58, 2015
|
|
7O48
 
 | |
4QST
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with 1-methylquinolin 2-one | | Descriptor: | 1,2-ETHANEDIOL, 1-METHYLQUINOLIN-2(1H)-ONE, ATPase family AAA domain-containing protein 2, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
7O2O
 
 | |
5JPZ
 
 | | Crystal structure of HAT domain of human Squamous Cell Carcinoma Antigen Recognized By T Cells 3, SART3 (TIP110) | | Descriptor: | Squamous cell carcinoma antigen recognized by T-cells 3 | | Authors: | Grazette, A, Harper, S, Emsley, J, Layfield, R, Dreveny, I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.045 Å) | | Cite: | unpublished
To Be Published
|
|
7O3D
 
 | | Cooperation between the intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly | | Descriptor: | Transcription elongation factor SPT6 | | Authors: | Kasiliauskaite, A, Kubicek, K, Klumpler, T, Zanova, M, Zapletal, D, Novacek, J, Stefl, R. | | Deposit date: | 2021-04-01 | | Release date: | 2022-04-13 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Nucleic Acids Res., 50, 2022
|
|
