6N7T
 
 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form III) | | Descriptor: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N7N
 
 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form I) | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N7S
 
 | | Structure of bacteriophage T7 E343Q mutant gp4 helicase-primase in complex with ssDNA, dTTP, AC dinucleotide and CTP (form II) | | Descriptor: | DNA (25-MER), DNA primase/helicase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
6N9W
 
 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS2) | | Descriptor: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Cui, Y, Zhou, Z, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
4DF3
 
 | |
2D2D
 
 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor I2 | | Descriptor: | 3C-like proteinase, ETHYL (2E,4S)-4-[((2R)-2-{[N-(TERT-BUTOXYCARBONYL)-L-VALYL]AMINO}-2-PHENYLETHANOYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENT-2-ENOATE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Ding, Y, Rao, Z. | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
9LTK
 
 | | Crystal structur of Lpg1618(R215F) from Legionella pneumophila | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Liu, T, Gao, J, Ge, H. | | Deposit date: | 2025-02-06 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and molecular characterization of AmpS, a class D beta-lactamase from Legionella pneumophila.
Int.J.Biol.Macromol., 312, 2025
|
|
3KRJ
 
 | | cFMS tyrosine kinase in complex with 4-Cyano-1H-imidazole-2-carboxylic acid (2-cyclohex-1-enyl-4-piperidin-4-yl-phenyl)-amide | | Descriptor: | 4-cyano-N-(2-cyclohex-1-en-1-yl-4-piperidin-4-ylphenyl)-1H-imidazole-2-carboxamide, ACETATE ION, Macrophage colony-stimulating factor 1 receptor, ... | | Authors: | Schubert, C. | | Deposit date: | 2009-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Optimization of a Potent Class of Arylamide Colony-Stimulating Factor-1 Receptor Inhibitors Leading to Anti-inflammatory Clinical Candidate 4-Cyano-N-[2-(1-cyclohexen-1-yl)-4-[1-[(dimethylamino)acetyl]-4-piperidinyl]phenyl]-1H-imidazole-2-carboxamide (JNJ-28312141).
J.Med.Chem., 54, 2011
|
|
3KRL
 
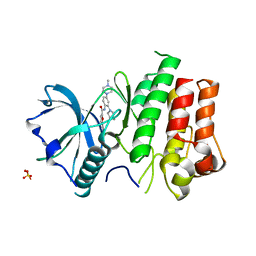 | | cFMS Tyrosine kinase in complex with 5-Cyano-furan-2-carboxylic acid [4-(4-methyl-piperazin-1-yl)-2-piperidin-1-yl-phenyl]-amide | | Descriptor: | 5-cyano-N-[4-(4-methylpiperazin-1-yl)-2-piperidin-1-ylphenyl]furan-2-carboxamide, Macrophage colony-stimulating factor 1 receptor, Basic fibroblast growth factor receptor 1, ... | | Authors: | Schubert, C. | | Deposit date: | 2009-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of a Potent Class of Arylamide Colony-Stimulating Factor-1 Receptor Inhibitors Leading to Anti-inflammatory Clinical Candidate 4-Cyano-N-[2-(1-cyclohexen-1-yl)-4-[1-[(dimethylamino)acetyl]-4-piperidinyl]phenyl]-1H-imidazole-2-carboxamide (JNJ-28312141).
J.Med.Chem., 54, 2011
|
|
1LCL
 
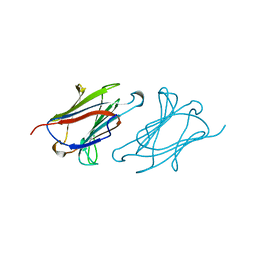 | | CHARCOT-LEYDEN CRYSTAL PROTEIN | | Descriptor: | LYSOPHOSPHOLIPASE | | Authors: | Acharya, K.R, Leonidas, D.D. | | Deposit date: | 1996-01-18 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Charcot-Leyden crystal protein, an eosinophil lysophospholipase, identifies it as a new member of the carbohydrate-binding family of galectins.
Structure, 3, 1995
|
|
2HNK
 
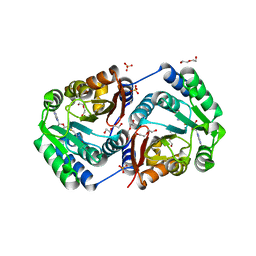 | | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent O-methyltransferase, ... | | Authors: | Hou, X, Wei, Z, Gong, W. | | Deposit date: | 2006-07-13 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SAM-dependent O-methyltransferase from pathogenic bacterium Leptospira interrogans.
J.Struct.Biol., 159, 2007
|
|
6UZB
 
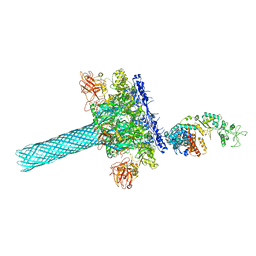 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6UZD
 
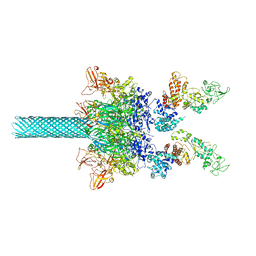 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
4JQH
 
 | |
6UZE
 
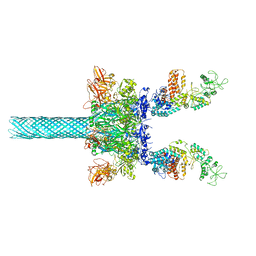 | | Anthrax toxin protective antigen channels bound to edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
6R1L
 
 | |
3DPK
 
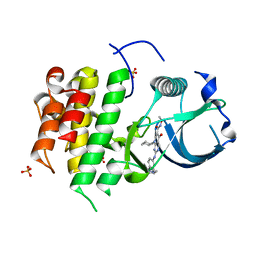 | | cFMS tyrosine kinase in complex with a pyridopyrimidinone inhibitor | | Descriptor: | 8-cyclohexyl-N-methoxy-5-oxo-2-{[4-(2-pyrrolidin-1-ylethyl)phenyl]amino}-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, Macrophage colony-stimulating factor 1 receptor, Fibroblast growth factor receptor 1, ... | | Authors: | Schubert, C. | | Deposit date: | 2008-07-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrido[2,3-d]pyrimidin-5-ones: a novel class of antiinflammatory macrophage colony-stimulating factor-1 receptor inhibitors
J.Med.Chem., 52, 2009
|
|
2LW6
 
 | | Solution structure of an avirulence protein AvrPiz-t from pathogen Magnaportheoryzae | | Descriptor: | AvrPiz-t protein | | Authors: | Zhang, Z.-M, Zhang, X, Zhou, Z, Hu, H, Liu, M, Zhou, B, Zhou, J. | | Deposit date: | 2012-07-23 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t.
J.Biomol.Nmr, 55, 2013
|
|
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
6AKL
 
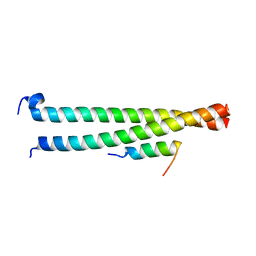 | | Crystal structure of Striatin3 in complex with SIKE1 Coiled-coil domain | | Descriptor: | Striatin-3, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
2KLZ
 
 | |
2JNH
 
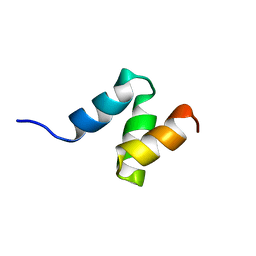 | | Solution Structure of the UBA Domain from Cbl-b | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Zhou, C, Zhou, Z, Lin, D, Hu, H. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Protein Sci., 17, 2008
|
|
6AKM
 
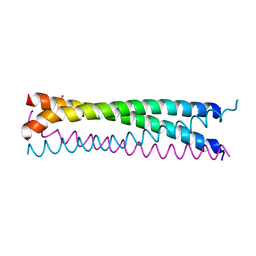 | | Crystal structure of SLMAP-SIKE1 complex | | Descriptor: | GLYCEROL, Sarcolemmal membrane-associated protein, Suppressor of IKBKE 1 | | Authors: | Ma, J, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
8E34
 
 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
4NZW
 
 | | Crystal Structure of STK25-MO25 Complex | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | Authors: | Feng, M, Hao, Q, Zhou, Z.C. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
