6CJ8
 
 | | CSP1 | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COQ
 
 | | CSP1-K6A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COP
 
 | | CSP1-R3A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COV
 
 | | CSP2-l14 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6COW
 
 | | CSP1 | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
7EYI
 
 | |
6DCB
 
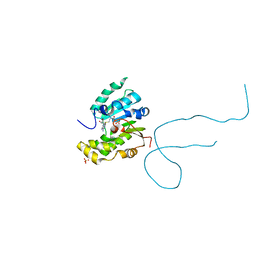 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
6DCC
 
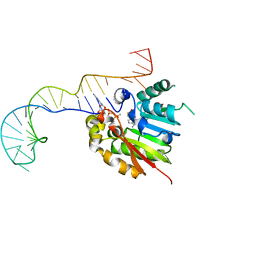 | | Structure of methylphosphate capping enzyme methyltransferase domain in complex with 5' end of 7SK RNA | | Descriptor: | 7SK snRNA methylphosphate capping enzyme, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Yang, Y, Eichhorn, C, Cascio, D, Feigon, J. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of 7SK RNA 5'-gamma-phosphate methylation and retention by MePCE.
Nat. Chem. Biol., 15, 2019
|
|
2JZ0
 
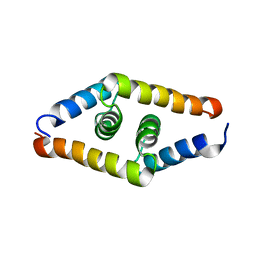 | | DSX_short | | Descriptor: | Protein doublesex | | Authors: | Yang, Y, Zhang, W, Bayrer, J.R, Weiss, M.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
J.Biol.Chem., 283, 2008
|
|
2JZ1
 
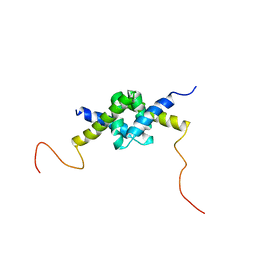 | | DSX_long | | Descriptor: | Protein doublesex | | Authors: | Yang, Y, Zhang, W, Bayrer, J.R, Weiss, M.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
J.Biol.Chem., 283, 2008
|
|
2KMM
 
 | | Solution NMR structure of the TGS domain of PG1808 from Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target PgR122A (418-481) | | Descriptor: | Guanosine-3',5'-bis(Diphosphate) 3'-pyrophosphohydrolase | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the TGS domain of PG1808 from
Porphyromonas gingivalis. Northeast Structural Genomics Consortium Target
PgR122A (418-481)
To be Published
|
|
6BVP
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6CGF
 
 | | Crystal structure of HIV-1 Y188L mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6CD3
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans in complex with 3-HAA | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6D62
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6C0N
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6BVQ
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase N27A from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6C0O
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6BVR
 
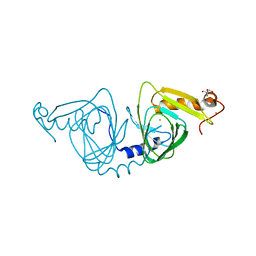 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142A from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2017-12-13 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6C0L
 
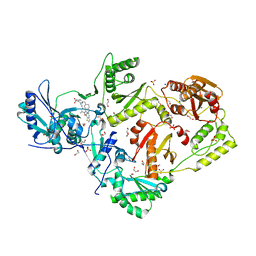 | | Crystal structure of HIV-1 E138K mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C0J
 
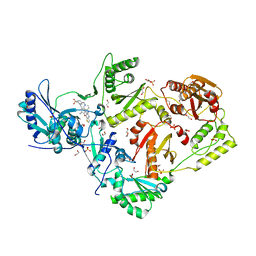 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6D60
 
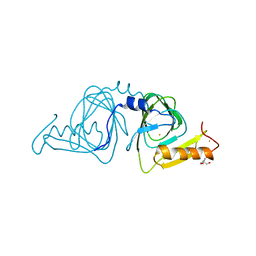 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6C0P
 
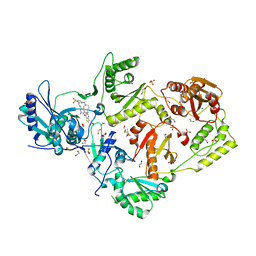 | | Crystal structure of HIV-1 E138K mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6DUG
 
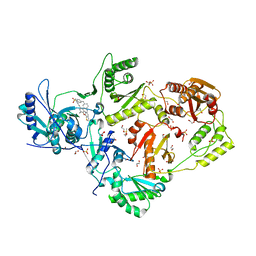 | | Crystal structure of HIV-1 reverse transcriptase K101P mutant in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-06-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.225 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6D61
 
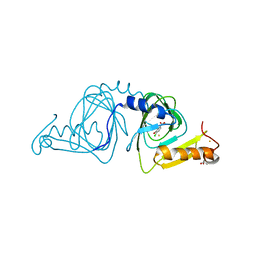 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
