6AJB
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADH, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
6AJ6
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADP+ | | Descriptor: | Isocitrate dehydrogenase [NADP], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
7X39
 
 | | Structure of CIZ1 bound ERH | | Descriptor: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
6AJ8
 
 | | Crystal structure of Trypanosoma brucei glycosomal isocitrate dehydrogenase in complex with NADP+, alpha-ketoglutarate and ca2+ | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Wang, X, Inaoka, D.K, Shiba, T, Balogun, E.O, Ziebart, N, Allman, S, Watanabe, Y, Nozaki, T, Boshart, M, Bringaud, F, Harada, S, Kita, K. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biochemical characterization of a novel Trypanosoma brucei glycosomal isocitrate dehydrogenase with dual coenzyme specificity (NADP+/NAD+)
To Be Published
|
|
8GX9
 
 | |
8HXK
 
 | | BANAL-20-236 S1 in complex with R. Affinis ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X, Xu, G. | | Deposit date: | 2023-01-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The selective effect of fecal-oral transmission on the S proteins of bat SARS-CoV-2 related coronaviruses in favor of stability over infectivity
To Be Published
|
|
8HXJ
 
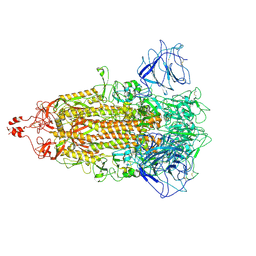 | | BANAL-20-52 Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Xu, G. | | Deposit date: | 2023-01-04 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The selective effect of fecal-oral transmission on the S proteins of bat SARS-CoV-2 related coronaviruses in favor of stability over infectivity
To Be Published
|
|
8I3W
 
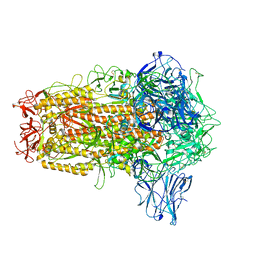 | | BANAL-20-236 Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Xu, G. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The selective effect of fecal-oral transmission on the S proteins of bat SARS-CoV-2 related coronaviruses in favor of stability over infectivity
To Be Published
|
|
2JT3
 
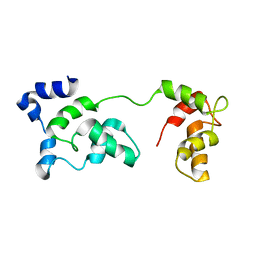 | | Solution Structure of F153W cardiac troponin C | | Descriptor: | Troponin C | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JTZ
 
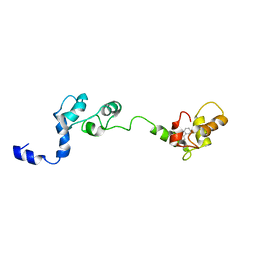 | | Solution structure and chemical shift assignments of the F104-to-5-flurotryptophan mutant of cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
2JZC
 
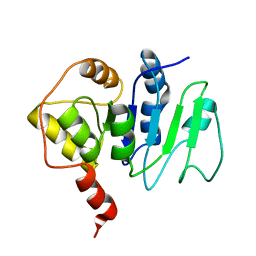 | | NMR solution structure of ALG13: The sugar donor subunit of a yeast N-acetylglucosamine transferase. Northeast Structural Genomics Consortium target YG1 | | Descriptor: | UDP-N-acetylglucosamine transferase subunit ALG13 | | Authors: | Wang, X, Weldeghorghis, T, Zhang, G, Imepriali, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase.
Structure, 16, 2008
|
|
2GMG
 
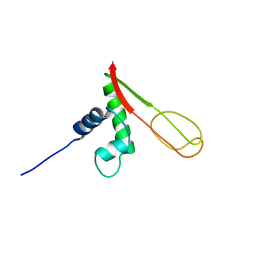 | | Solution NMR Structure of protein PF0610 from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfG3 | | Descriptor: | hypothetical protein Pf0610 | | Authors: | Wang, X, Lee, H.S, Adams, M.W, Northeast Structural Genomics Consortium (NESG), Montelione, G.T, Prestegard, J.H. | | Deposit date: | 2006-04-06 | | Release date: | 2006-11-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PF0610, a novel winged helix-turn-helix variant possessing a rubredoxin-like Zn ribbon motif from the hyperthermophilic archaeon, Pyrococcus furiosus.
Biochemistry, 46, 2007
|
|
2K2R
 
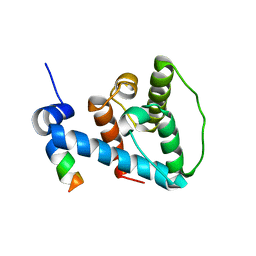 | | The NMR structure of alpha-parvin CH2/paxillin LD1 complex | | Descriptor: | Alpha-parvin, Paxillin | | Authors: | Wang, X, Fukuda, K, Byeon, I, Velyvis, A, Wu, C, Gronenborn, A, Qin, J. | | Deposit date: | 2008-04-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
J.Biol.Chem., 283, 2008
|
|
2JT0
 
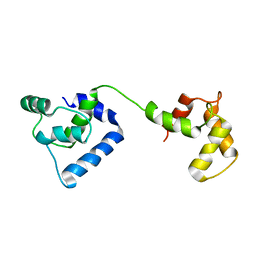 | | Solution structure of F104W cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P.-J, Sykes, B.D. | | Deposit date: | 2007-07-17 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JT8
 
 | | Solution structure of the F153-to-5-flurotryptophan mutant of human cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
7CYH
 
 | |
7EP9
 
 | |
7DH6
 
 | | Crystal structure of PLRG1 | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Pleiotropic regulator 1, ... | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2020-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Crystal structure of the WD40 domain of human PLRG1.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
5J8V
 
 | |
8ZL9
 
 | | ASFV p72 in complex with Fab G6 | | Descriptor: | B646L, G6 Heavy chain, G6 Light chain | | Authors: | Wang, X, Fu, W, Yu, Q. | | Deposit date: | 2024-05-17 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | p72 antigenic mapping reveals a potential supersite of vulnerability for African swine fever virus.
Cell Discov, 10, 2024
|
|
8E83
 
 | |
8EA2
 
 | |
8EA1
 
 | |
9CKV
 
 | | Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Werther, R, Nguyen, A, Estrada Alamo, K.A, Wang, X, Campbell, M.G. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | De Novo Design of Integrin alpha5beta1 Modulating Proteins for Regenerative Medicine
To Be Published
|
|
7BWJ
 
 | | crystal structure of SARS-CoV-2 antibody with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Wang, X, Ge, J. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human neutralizing antibodies elicited by SARS-CoV-2 infection.
Nature, 584, 2020
|
|
