6NET
 
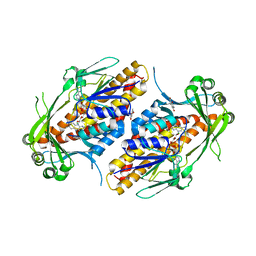 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | Descriptor: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NKK
 
 | | Structure of PhqE Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and premalbrancheamide | | Descriptor: | (5aS,12aS,13aS)-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6NEU
 
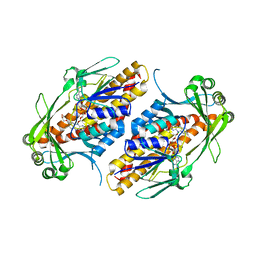 | | FAD-dependent monooxygenase TropB from T. stipitatus R206Q variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NKM
 
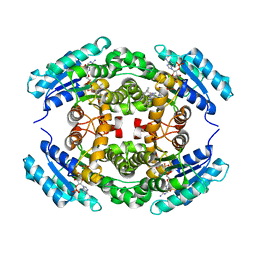 | | Structure of PhqE D166N Reductase/Diels-Alderase from Penicillium fellutanum in complex with NADP+ and substrate | | Descriptor: | 3-{[2-(2-methylbut-3-en-2-yl)-1H-indol-3-yl]methyl}-8H-pyrrolo[1,2-a]pyrazin-5-ium-1-olate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short chain dehydrogenase | | Authors: | Newmister, S.A, Dan, Q, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
6NEV
 
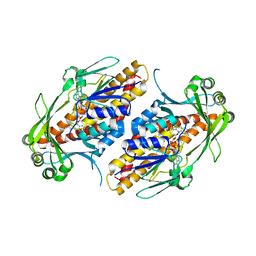 | | FAD-dependent monooxygenase TropB from T. stipitatus Y239F Variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
8SY2
 
 | |
6NES
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NKH
 
 | |
6NKI
 
 | | Structure of PhqB Reductase Domain from Penicillium fellutanum | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NRPS | | Authors: | Dan, Q, Newmister, S.A, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-01-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase.
Nat.Chem., 11, 2019
|
|
5DOZ
 
 | | Crystal structure of JamJ enoyl reductase (NADPH bound) | | Descriptor: | ACETATE ION, JamJ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
5DOV
 
 | |
1O57
 
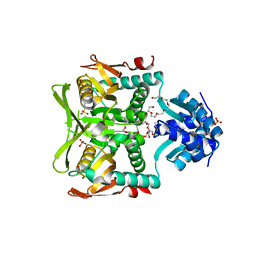 | | CRYSTAL STRUCTURE OF THE PURINE OPERON REPRESSOR OF BACILLUS SUBTILIS | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Sinha, S.C, Krahn, J, Shin, B.S, Tomchick, D.R, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Purine Repressor of Bacillus Subtilis: A Novel Combination of Domains Adapted for Transcription Regulation
J.Bacteriol., 185, 2003
|
|
1NON
 
 | | PyrR, the regulator of the pyrimidine biosynthetic operon in Bacillus caldolyticus | | Descriptor: | PyrR bifunctional protein | | Authors: | Switzer, R.L, Chander, P, Smith, J.L, Halbig, K.M, Miller, J.K, Bonner, H.K, Grabner, G.K. | | Deposit date: | 2003-01-16 | | Release date: | 2004-05-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides
J.Bacteriol., 187, 2005
|
|
5DP2
 
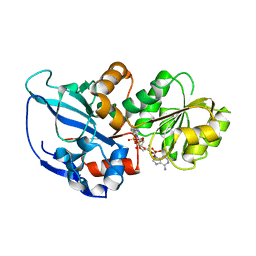 | |
5DP1
 
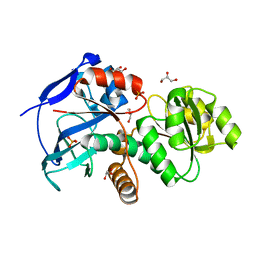 | | Crystal structure of CurK enoyl reductase | | Descriptor: | CurK, GLYCEROL, PHOSPHATE ION | | Authors: | Khare, D, Smith, J.L. | | Deposit date: | 2015-09-12 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Cyclopropanation by a Unique Enoyl-Acyl Carrier Protein Reductase.
Structure, 23, 2015
|
|
1P4A
 
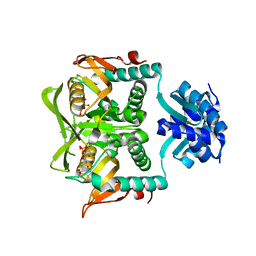 | | Crystal Structure of the PurR complexed with cPRPP | | Descriptor: | 1-ALPHA-PYROPHOSPHORYL-2-ALPHA,3-ALPHA-DIHYDROXY-4-BETA-CYCLOPENTANE-METHANOL-5-PHOSPHATE, Pur operon repressor | | Authors: | Bera, A.K, Zhu, J, Zalkin, H, Smith, J.L. | | Deposit date: | 2003-04-22 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Functional dissection of the Bacillus subtilis pur operator site.
J.Bacteriol., 185, 2003
|
|
1ECJ
 
 | |
6D5X
 
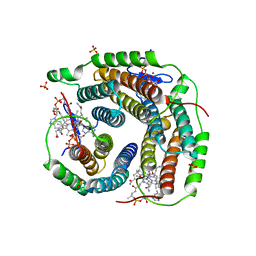 | | Structure of Human ATP:Cobalamin Adenosyltransferase bound to ATP, Adenosylcobalamin, and Triphosphate | | Descriptor: | 5'-DEOXYADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, COBALAMIN, ... | | Authors: | Dodge, G.J, Campanello, G, Smith, J.L, Banerjee, R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12as a Cofactor Conservation Strategy.
J. Am. Chem. Soc., 140, 2018
|
|
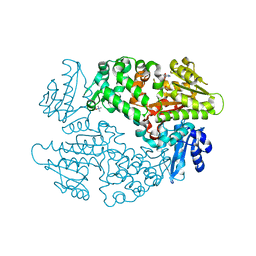
6D6Y
 
 | |
6D5K
 
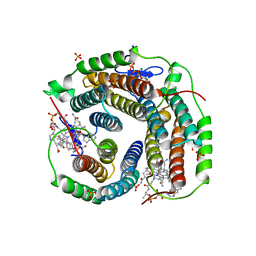 | | Structure of Human ATP:Cobalamin Adenosyltransferase bound to ATP, and Adenosylcobalamin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXYADENOSINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dodge, G.J, Campanello, G, Smith, J.L, Banerjee, R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Sacrificial Cobalt-Carbon Bond Homolysis in Coenzyme B12as a Cofactor Conservation Strategy.
J. Am. Chem. Soc., 140, 2018
|
|
9E50
 
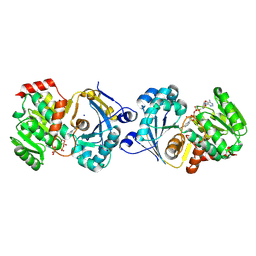 | |
6ECW
 
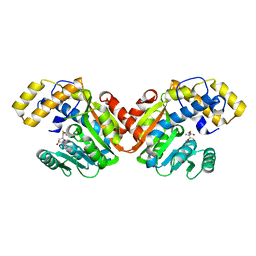 | | StiD O-MT residues 956-1266 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECX
 
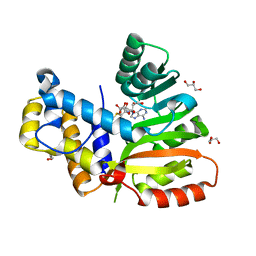 | | StiE O-MT residues 942-1257 | | Descriptor: | GLYCEROL, S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.B, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECV
 
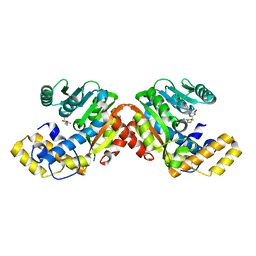 | | StiD O-MT residues 976-1266 | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, StiD protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
6ECT
 
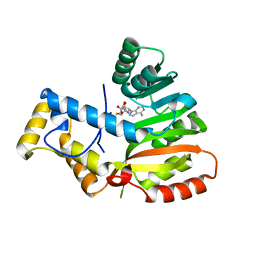 | | StiE O-MT residues 961-1257 | | Descriptor: | S-ADENOSYLMETHIONINE, StiE protein | | Authors: | Skiba, M.A, Bivins, M.M, Smith, J.L. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis of Polyketide Synthase O-Methylation.
ACS Chem. Biol., 13, 2018
|
|
