8AUI
 
 | |
8AUJ
 
 | |
8AUL
 
 | |
8AUM
 
 | |
8AU9
 
 | |
8AUA
 
 | |
8AUF
 
 | |
8AUN
 
 | |
8AUO
 
 | |
8AUE
 
 | |
8AUB
 
 | |
8AUQ
 
 | |
6WW8
 
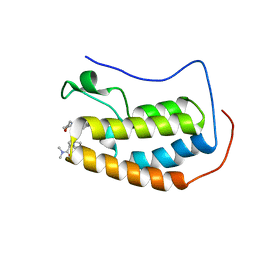 | | BRD4 Bromodomain 1 in complex with triple CDK4/6-PI3K-BET inhibitor | | Descriptor: | 7-cyclopentyl-N,N-dimethyl-2-({4-[5-(morpholin-4-yl)-7-oxo-7H-thieno[3,2-b]pyran-3-yl]phenyl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Vann, K.R, Kutateladze, T.G. | | Deposit date: | 2020-05-08 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | A triple action CDK4/6-PI3K-BET inhibitor with augmented cancer cell cytotoxicity.
Cell Discov, 6, 2020
|
|
8CUH
 
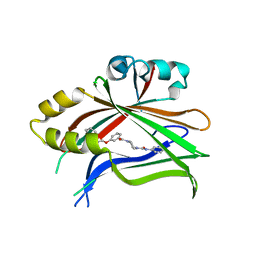 | | Crystal structure of human TEAD2 complexed with its inhibitor TM2. | | Descriptor: | 4-[3-(2-cyclohexylethoxy)benzoyl]-N-phenylpiperazine-1-carboxamide, Transcriptional enhancer factor TEF-4 | | Authors: | Liu, S, Luo, X. | | Deposit date: | 2022-05-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a new class of reversible TEA-domain transcription factor inhibitors with a novel binding mode.
Elife, 11, 2022
|
|
6YMV
 
 | |
6YMW
 
 | |
9C93
 
 | | Crystal structure of menin in complex with inhibitor compound 26 | | Descriptor: | (1R,5R)-2-({5-fluoro-2-[(5-{7-[(1-methylcyclopropyl)methyl]-2,7-diazaspiro[3.5]nonan-2-yl}-1,2,4-triazin-6-yl)oxy]phenyl}methyl)-2-azabicyclo[3.1.0]hexan-3-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Menin, ... | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Potent Menin-KMT2A Interaction Inhibitors with Improved In Vitro ADME Properties and Reduced hERG Affinity.
Acs Med.Chem.Lett., 16, 2025
|
|
9C92
 
 | | Crystal structure of menin in complex with inhibitor compound 15 | | Descriptor: | Menin, N-ethyl-5-fluoro-2-[(5-{7-[(1-methylcyclopropyl)methyl]-2,7-diazaspiro[3.5]nonan-2-yl}-1,2,4-triazin-6-yl)oxy]-N-(propan-2-yl)benzamide | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Potent Menin-KMT2A Interaction Inhibitors with Improved In Vitro ADME Properties and Reduced hERG Affinity.
Acs Med.Chem.Lett., 16, 2025
|
|
9C94
 
 | | Crystal structure of menin in complex with inhibitor compound 20 | | Descriptor: | Menin, {5-fluoro-2-[(5-{7-[(1-methylcyclopropyl)methyl]-2,7-diazaspiro[3.5]nonan-2-yl}-1,2,4-triazin-6-yl)oxy]phenyl}[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Bester, S.M, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-06-13 | | Release date: | 2025-01-15 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design of Potent Menin-KMT2A Interaction Inhibitors with Improved In Vitro ADME Properties and Reduced hERG Affinity.
Acs Med.Chem.Lett., 16, 2025
|
|
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EDF
 
 | |
8ECV
 
 | | Bovine Fab 2F12 | | Descriptor: | 2F12 Fab Heavy chain, 2F12 Fab Light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | Descriptor: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7LNK
 
 | |
