1E57
 
 | |
1GCA
 
 | |
6M1W
 
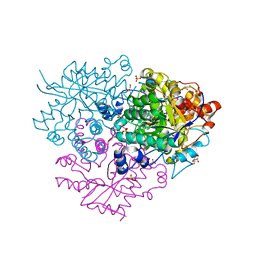 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | Amidohydrolase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
6M2I
 
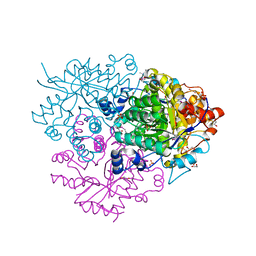 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
7QWL
 
 | |
7QVC
 
 | |
7QVF
 
 | |
7QWG
 
 | |
7QWM
 
 | |
1URP
 
 | |
1DBP
 
 | |
2MGO
 
 | | NMR solution structure of oxytocin | | Descriptor: | Oxytocin | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2013-11-01 | | Release date: | 2014-10-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M9O
 
 | | Solution structure of kalata B7 | | Descriptor: | Kalata-B7 | | Authors: | Daly, N, Elliott, A, Craik, D. | | Deposit date: | 2013-06-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cyclotides as templates for peptide GPCR ligand design - discovery of the target receptor of the oxytocic plant peptide kalata B7
Proc.Natl.Acad.Sci.USA, 2013
|
|
1DRK
 
 | |
4C8B
 
 | | Structure of the kinase domain of human RIPK2 in complex with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Canning, P, Krojer, T, Bradley, A, Mahajan, P, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Inflammatory Signaling by NOD-RIPK2 Is Inhibited by Clinically Relevant Type II Kinase Inhibitors.
Chem. Biol., 22, 2015
|
|
1DRJ
 
 | |
6IYI
 
 | |
6IYH
 
 | |
1AMX
 
 | |
2PH6
 
 | | Crystal Structure of Human Beta Secretase Complexed with inhibitor | | Descriptor: | 3-({[(1R)-1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)-5-[METHYL(METHYLSULFONYL)AMINO]BENZYL ALPHA-METHYL-D-PHENYLALANINATE, Beta-secretase 1, SULFATE ION | | Authors: | Munshi, S. | | Deposit date: | 2007-04-10 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and SAR of macrocyclic tertiary carbinamine BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7ST4
 
 | | Calcium-saturated jGCaMP8.410.80 | | Descriptor: | CALCIUM ION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Zhang, Y, Looger, L.L. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fast and sensitive GCaMP calcium indicators for imaging neural populations
Nature, 615, 2023
|
|
5QCW
 
 | | Crystal structure of BACE complex with BMC021 | | Descriptor: | (2R,4S)-N-butyl-4-[(2S,5S,7R)-2,7-dimethyl-3,15-dioxo-1,4-diazacyclopentadecan-5-yl]-4-hydroxy-2-methylbutanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QD7
 
 | | Crystal structure of BACE complex with BMC014 | | Descriptor: | (4S)-4-[(1R)-1-hydroxy-2-({1-[3-(1-methylethyl)phenyl]cyclopropyl}amino)ethyl]-19-(methoxymethyl)-11-oxa-3,16-diazatric yclo[15.3.1.1~6,10~]docosa-1(21),6(22),7,9,17,19-hexaen-2-one, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
5QDB
 
 | | Crystal structure of BACE complex with BMC002 | | Descriptor: | (3S,14R,16S)-16-[(1R)-1-hydroxy-2-{[3-(1-methylethyl)benzyl]amino}ethyl]-3,4,14-trimethyl-1,4-diazacyclohexadecane-2,5- dione, Beta-secretase 1 | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
2PH8
 
 | | Crystal Structure of Human Beta Secretase Complexed with inhibitor | | Descriptor: | Beta-secretase 1, N-[(5R,14R)-5-AMINO-5,14-DIMETHYL-4-OXO-3-OXA-18-AZATRICYCLO[15.3.1.1~7,11~]DOCOSA-1(21),7(22),8,10,17,19-HEXAEN-19-YL]-N-METHYLMETHANESULFONAMIDE, SULFATE ION | | Authors: | Munshi, S. | | Deposit date: | 2007-04-10 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Strategies toward improving the brain penetration of macrocyclic tertiary carbinamine BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
