5DBI
 
 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ and 10-oxogeranial | | Descriptor: | (2E,6E)-2,6-dimethylocta-2,6-dienedial, Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5D4Y
 
 | | A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, xylanase | | Authors: | Zheng, Y, Guo, R.T. | | Deposit date: | 2015-08-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into potential cold adaptation mechanism through a psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase.
J.Struct.Biol., 193, 2016
|
|
5DBG
 
 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NAD+ | | Descriptor: | Iridoid synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Liu, W.D, Hu, Y.M, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6JJS
 
 | | Crystal structure of an enzyme from Penicillium herquei in condition2 | | Descriptor: | ACETATE ION, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
6JJT
 
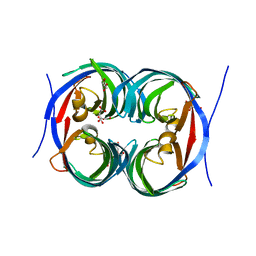 | | Crystal structure of an enzyme from Penicillium herquei in condition1 | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, PhnH | | Authors: | Fen, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-02-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure and proposed mechanism of an enantioselective hydroalkoxylation enzyme from Penicillium herquei.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
5CUY
 
 | | Crystal structure of Trypanosoma brucei Vacuolar Soluble Pyrophosphatases in apo form | | Descriptor: | Acidocalcisomal pyrophosphatase, CITRIC ACID, MAGNESIUM ION | | Authors: | Yang, Y.Y, Ko, T.P, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
6ITA
 
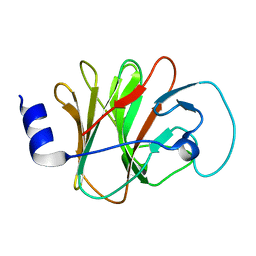 | | Crystal structure of intracellular B30.2 domain of BTN3A1 mutant | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-11-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5DBF
 
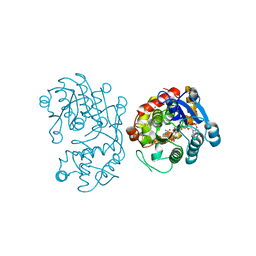 | | Crystal Structure of Iridoid Synthase from Cantharanthus roseus in complex with NADPH | | Descriptor: | Iridoid synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hu, Y.M, Liu, W.D, Zheng, Y.Y, Xu, Z.X, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-08-21 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Iridoid Synthase from Cantharanthus roseus with Bound NAD(+) , NADPH, or NAD(+) /10-Oxogeranial: Reaction Mechanisms
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5EPT
 
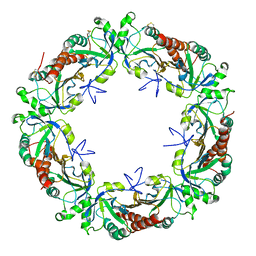 | |
5CUV
 
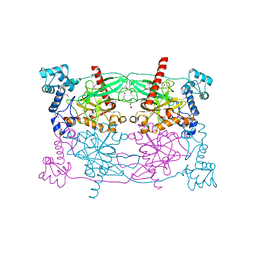 | | Crystal structure of Trypanosoma cruzi Vacuolar Soluble Pyrophosphatases in apo form | | Descriptor: | Acidocalcisomal pyrophosphatase, D-MALATE, MAGNESIUM ION | | Authors: | Ko, T.P, Yang, Y.Y, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2015-07-25 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of Trypanosoma cruzi protein in complex with ligand
Acs Chem.Biol., 2016
|
|
6KA2
 
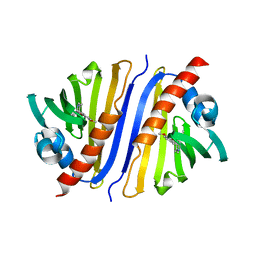 | | Crystal structure of a Thebaine synthase from Papaver somniferum in complex with TBN | | Descriptor: | (4R,7aR,12bS)-7,9-dimethoxy-3-methyl-2,4,7a,13-tetrahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline, Thebaine synthase 2 | | Authors: | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
5DVB
 
 | |
5CUW
 
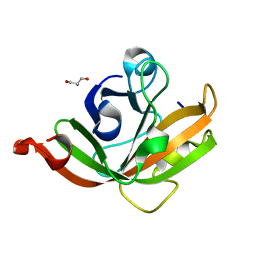 | |
6KA3
 
 | | Crystal structure of a Thebaine synthase from Papaver somniferum | | Descriptor: | PALMITIC ACID, SULFATE ION, Thebaine synthase 2 | | Authors: | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
6KQX
 
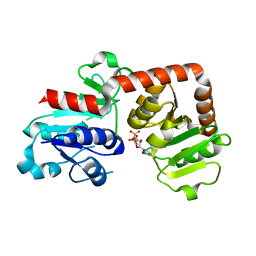 | | Crystal structure of Yijc from B. subtilis in complex with UDP | | Descriptor: | URIDINE-5'-DIPHOSPHATE, Uncharacterized UDP-glucosyltransferase YjiC | | Authors: | Hu, Y.M, Dai, L.H, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural dissection of unnatural ginsenoside-biosynthetic UDP-glycosyltransferase Bs-YjiC from Bacillus subtilis for substrate promiscuity.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
6A9F
 
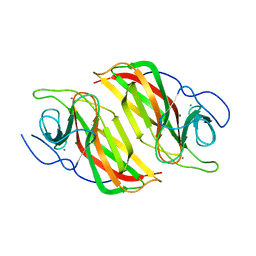 | | Crystal structure of a cyclase from Fischerella sp. TAU in complex with 4-(1H-Indol-3-yl)butan-2-one | | Descriptor: | 4-(1~{H}-indol-3-yl)butan-2-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-13 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7CWQ
 
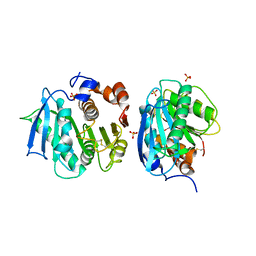 | | Crystal structure of a novel cutinase from Burkhoderiales bacterium RIFCSPLOWO2_02_FULL_57_36 | | Descriptor: | DLH domain-containing protein, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
7CQB
 
 | |
6B73
 
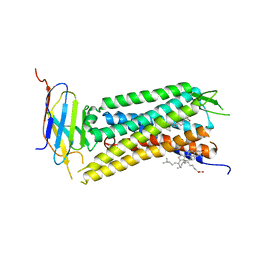 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
6CHR
 
 | | Crystal structure of a group II intron lariat with an intact 3' splice site (pre-2s state) | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*A)-3'), RNA (621-MER), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-22 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
6C8Y
 
 | | D30N HIV-1 protease in complex with a phenylboronic acid (P2') analog of darunavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Windsor, I.W, Raines, R.T, Forest, K.T. | | Deposit date: | 2018-01-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Sub-picomolar Inhibition of HIV-1 Protease with a Boronic Acid.
J. Am. Chem. Soc., 140, 2018
|
|
6BQC
 
 | | Cyclopropane fatty acid synthase from E. coli | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CARBONATE ION, Cyclopropane-fatty-acyl-phospholipid synthase, ... | | Authors: | Hari, S.B, Grant, R.A, Sauer, R.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.073 Å) | | Cite: | Structural and Functional Analysis of E. coli Cyclopropane Fatty Acid Synthase.
Structure, 26, 2018
|
|
6CIH
 
 | | Crystal structure of a group II intron lariat in the post-catalytic state | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, RNA (5'-R(P*UP*GP*UP*UP*UP*AP*UP*UP*AP*AP*AP*AP*AP*C*-3'), ... | | Authors: | Chan, R.T, Peters, J.K, Robart, A.R, Wiryaman, T, Rajashankar, K.R, Toor, N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.676 Å) | | Cite: | Structural basis for the second step of group II intron splicing.
Nat Commun, 9, 2018
|
|
7CY0
 
 | | Crystal structure of S185H mutant PET hydrolase from Ideonella sakaiensis | | Descriptor: | ACETIC ACID, Poly(ethylene terephthalate) hydrolase | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
6BRA
 
 | |
